Figure 4. Fibrillarin and RNA pol I form high concentration assemblies in the absence of rDNA.
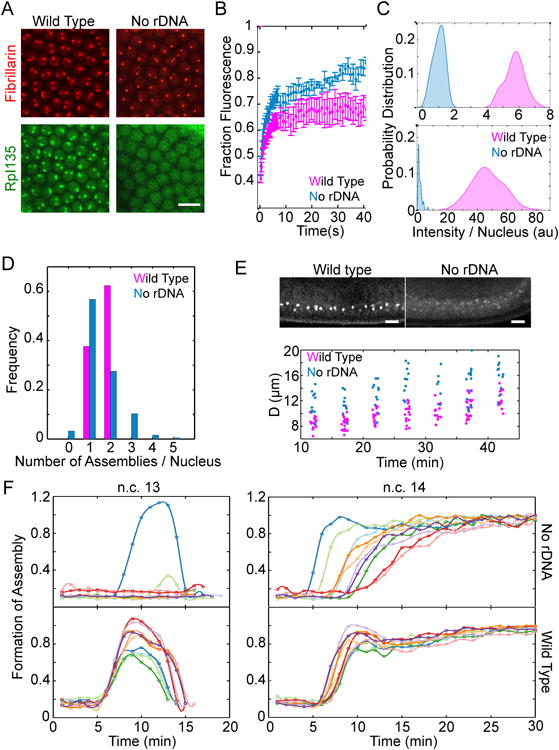
A. Representative images of the nuclei for wild type versus mutant embryos lacking rDNA at n.c. 14 show that both RFP-Fib and RpI135-GFP can form high concentration assemblies in the absence of rDNA. B. FRAP of Fibrillarin assemblies for the nucleoli and HANPs show that both are dynamic structures (rate constant for unbinding is 0.34 ± 0.03 s-1 for nucleoli and 0.27 ± 0.04 s-1 for HANPs). C. Histogram of the integrated intensity of nucleolus and HANPs normalized to the mean intensity of HANPs is shown for RFP-Fib (top) and RpI135-GFP (bottom). The probabilities for the case of RpI135-GFP in nucleolus is multiplied by 10 for better visualization. D. The fraction of nuclei showing different numbers of nucleoli in the wild-type or HANPs in the absence of rDNA is depicted for embryos after 15 minutes into n.c. 14. E. Top: Lateral view of the wild type and mutant embryos lacking rDNA at n.c. 14, expressing RFP-Fib. Bottom: quantification of the distance from the chorion of the nucleoli in a wild-type and HANPs in the mutant embryos lacking rDNA. F. Formation of fibrillarin high concentration assemblies for ten wild type (bottom) and ten mutant embryos lacking rDNA (top) over time at n.c. 13-14. Each color depicts an individual embryo. Time zero marks the end of mitosis. Scale bars 10 μm. See also Movie 2 and Figure S1.
