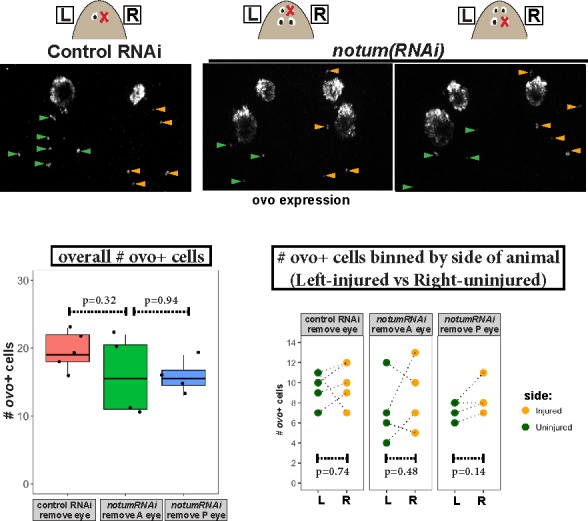
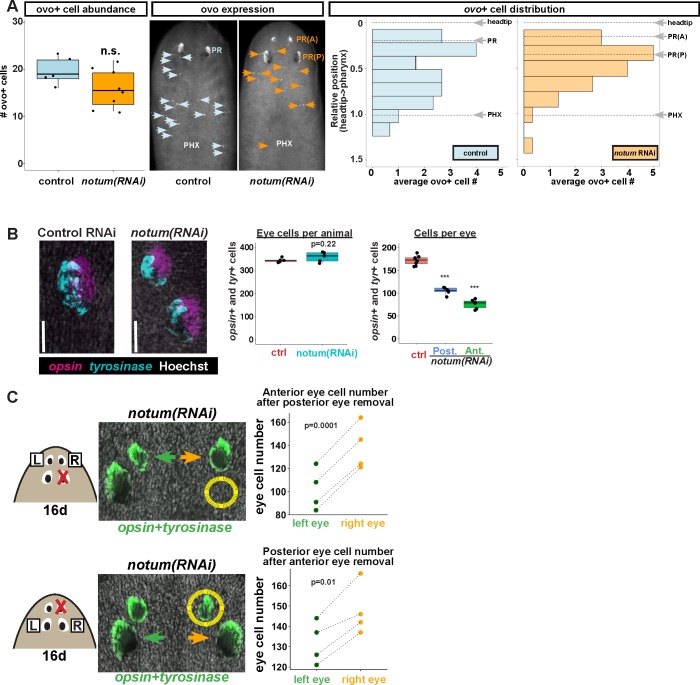
Figure 4. Non-regenerative eyes and regenerative eyes compete for progenitor acquisition.
(A) FISH to detect ovo+ progenitor cells located in the anterior animal region (middle panel, arrows) of control and notum(RNAi) animals. Left plots, ovo+ progenitor cell numbers were not significantly altered in 4-eyed notum(RNAi) animals. Right plots, histograms quantifying distribution of ovo+ eye cells showing regions anterior to the pharynx, with position normalized to the locations of the head tip and the pharynx. notum inhibition produced a slight anterior shift to the distribution of ovo+ cells, but they are present in a region that includes the posterior non-regenerative eyes. (B) FISH with opsin and tyrosinase riboprobes to detect numbers of eye cells from 4-eyed notum(RNAi) animals and 2-eyed control animals (bars, 25 microns). Hoechst counterstaining was used to count numbers of eye cells plotted below as total eye cell numbers per animal and cells per eye. notum RNAi did not significantly change total eye cell numbers, and reduced the number of cells per eye. Significance determined by 2-tailed t-test, ***p<0.001. (C) Four-eyed notum(RNAi) animals were generated by dsRNA feeding over 40 days prior to removal of either a posterior (top) or anterior (bottom) eye on one side of the animal (R,right), leaving both eyes on the left side (L) unaffected. After 16 days of recovery, animals were fixed and stained with a combination of riboprobes for opsin and tyrosinase (green), and eye cells were quantified by counting Hoechst-positive nuclei from opsin/tyrosinase +cells throughout the D/V eye axis. Right, quantifications of left and right eyes from several individuals are shown and connected by dotted lines. Top, removal of a posterior eye caused the ipsilateral anterior eye (orange) to become enlarged compared to the contralateral anterior eye (green). Bottom, removal of an anterior eye caused the ipsilateral posterior eye (orange) to become enlarged compared to the contralateral posterior eye (green). Significance was measured by 2-tailed paired sample t-tests.
Figure 4—figure supplement 1. Effect of nearby tissue removal on posterior eye regeneration ability in notum(RNAi) animals.
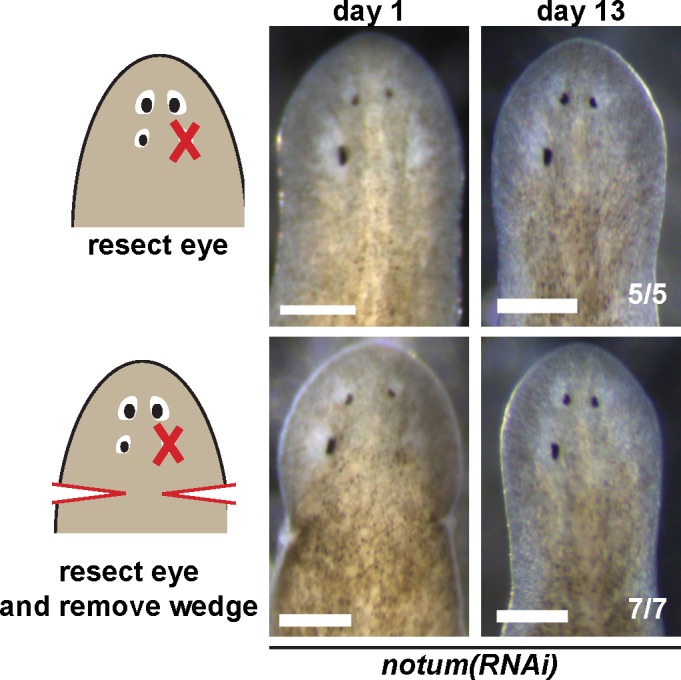
Figure 4—figure supplement 2. Measurement of ovo+ cell numbers after injury in control and notum(RNAi) animals.