Figure 2. The crystal structure of PtUGT1 with bound indoxyl sulfate.
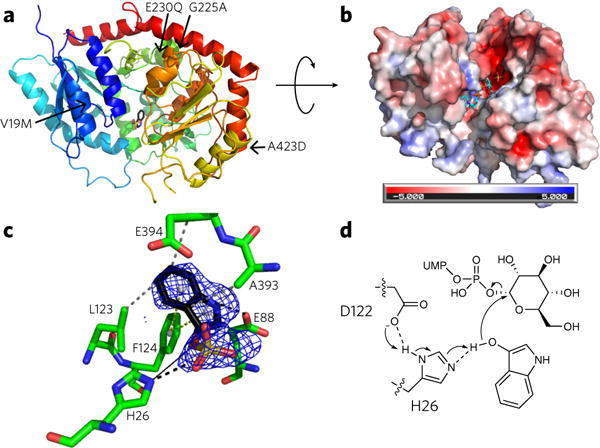
(a) Overall GT-B fold of PtUGT1 (PDB ID 5NLM). The N-terminal Rossmann domain (blue) consists of a seven-stranded parallel β-sheet surrounded by nine α-helical segments, and the C-terminal Rossmann domain (red) consists of a six-stranded parallel β-sheet and five α-helices. The indoxyl sulfate bound in the active site is shown in stick representation. The structure is annotated with the four amino acids differing between PtUGT1 and PtUGT2 (arrows). (b) Electrostatic surface potential of PtUGT1. Colors show the electrostatic potential, from −5 kT/e (red) to 5 kT/e (blue). The view is rotated ~90° around the x axis with respect to a. Bound indoxyl sulfate is depicted in stick representation (black) together with the donor substrate UDP-glucose superposed from the homologous AtUGT72B1 complex structure (cyan, PDB ID 2VCE). (c) The acceptor binding site with the bound indoxyl sulfate and interacting residues shown in stick representation. The indoxyl sulfate omit map is displayed as a blue grid contoured at 3.0 σ. Interactions are depicted as dashed lines (black, salt bridge/hydrogen bond; gray, hydrophobic interaction; yellow, π-stacking). A black solid line indicates the distance from the catalytic histidine to the glucose-accepting oxygen. (d) In the proposed catalytic mechanism based on homology to other characterized UGTs29,31, H26 deprotonates the indoxyl hydroxyl group, which then performs an SN2 attack on the anomeric carbon of glucose. The conserved D122 is believed to balance the charge on the catalytic histidine29. Consistent with this hypothesis, D122 forms a 2.6 Å hydrogen bond to H26 in the present structure.
