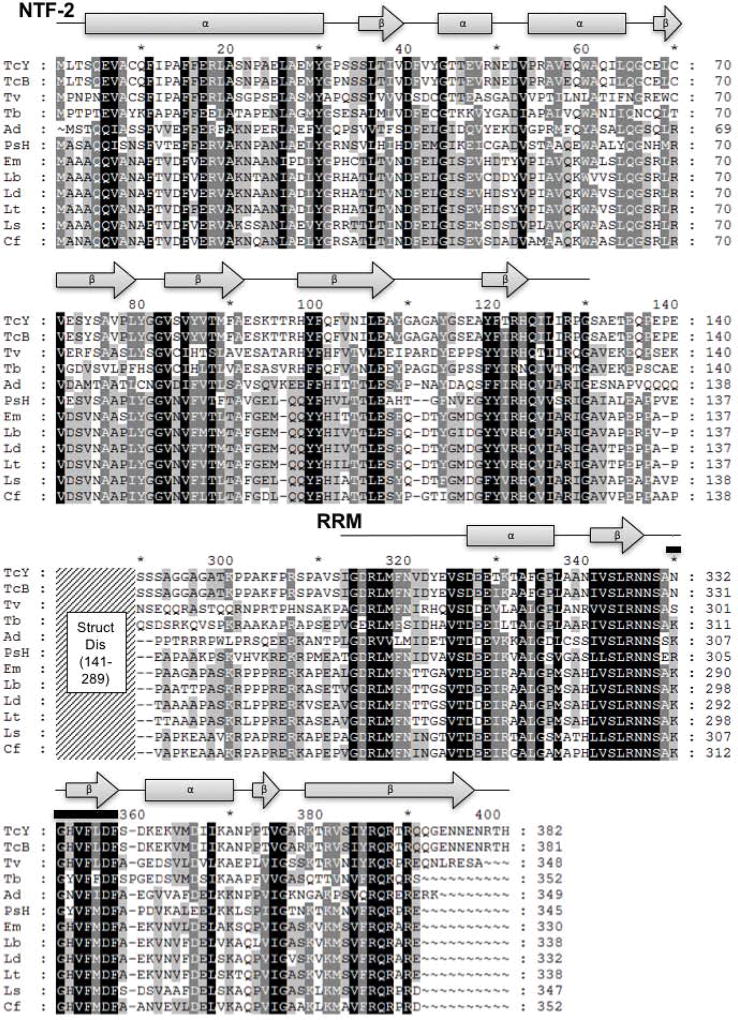
Figure 1. Kinetoplastid RBP42 homologues consist of an amino-terminal NTF2-like domain and a carboxy-terminal RRM domain.
Shown are the eight secondary structures that help define an NTF2-like domain and six secondary structures consistent with a RRM-like domain; filled gray rectangles represent predicted T. cruzi Y strain RBP42 α-helices, filled gray arrows represent predicted T. cruzi Y strain RBP42 β-strands, and black lines represent additional amino acids included in the domains. Within the T. cruzi Y strain RRM, the RNP1 RNA-binding signature sequence motif K/R-G-F/Y-G/A-F/Y-V/I/L-X-F/Y is found as NGHVFLDF (residues 350-357, amino acids are overlined); this sequence is highly conserved amongst the RBP42 homologues. The middle portion of the alignment, residues 141-289 represented by the diagonally-lined region, was omitted from the figure as this region is predicted to be structurally disordered (Struct Dis) and reveals less similarity among these sequences. Amino acid sequences are represented in single letter amino acid code; intensity of gray scale shading from black to no shading indicates degree of identity ranging from identical or highly similar to limited or no similarity. Secondary structure prediction based on the T. cruzi Y strain RBP42 amino acid sequence was done using the Phyre2 web portal (Kelley et al. 2015). I-TASSER, PSIPRED, and JPred all returned similar results to the Phyre2 prediction (Roy et al. 2012) (Drozdetskiy et al. 2015)
(Buchan et al. 2013) (Zhang 2009). Gene annotations are: TcY, T. cruzi Y strain; TcB, T. cruzi CL Brener Esmeraldo-like (TcCLB.509167.140); Tv, T. vivax Y486 (TvY486_0603830); Tb, T. brucei TREU927 (Tb927.6.4440); Ad, Angomonas deanei (gi: 528258120, gb: EPY37436.1); PsH, Phytomonas sp. isolate Hart1 (gi: 594148441, emb: CCW68155.1); Em, Endotrypanum monterogeii strain LV88 (EMOLV88_300036900); Lb, Leishmania braziliensis MHOM/BR/75/M2903 (LBRM2903_300037500); Ld, Leishmania donovani BPK282A1 (LdBPK_303130.1); Lt, Leishmania tropica L590 (LTRL590_300039500); Ls, Leptomonas seymouri ATCC 30220 (Lsey_0105_0110); Cf, Crithidia fasciculata strain Cf-Cl (CFAC1_260055900). NTF, nuclear transport factor; RRM, RNA recognition motif.