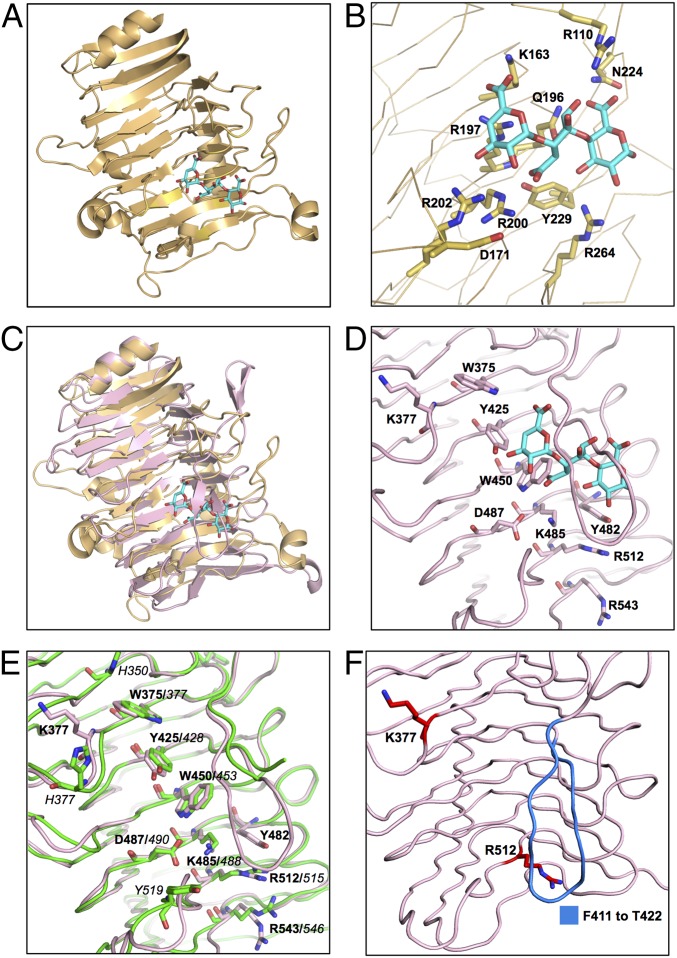
Fig. 3.
Comparison of the TM-Pel–binding pocket with the PuBS of SRRP53608-BR and SRRP100-23-BR. (A) Crystal structure of TM-Pel (Protein Data Bank ID code 3ZSC) (orange) in complex with TGA (cyan). (B) A close-up view of the TM-Pel–binding pocket, with residues involved in TGA binding represented as sticks. (C) Superposition of SRRP53608-BR262–571 (light pink) and TM-Pel (orange) structures, with an rmsd of 2.63 over 210 residues, shows that the PuBS of the former overlaps with that of TM-Pel. (D) Surface-exposed aromatic and charged residues on PB3 in SRRP53608-BR262–571 PuBS (light pink). This includes the aromatic residue triad W375, Y425, and W450 and the basic residue triad K485, R512, and R543. (E) Solvent-exposed residues of PuBS in the overlaid structures of SRRP53608-BR (light pink) and SRRP100-3-BR (green), with an rmsd of 0.912 over 293 residues, showing that several solvent-exposed residues in both binding sites are conserved. Residues in bold are from SRRP53608-BR, and those in italic font are from SRRP100-23-BR. (F) Residues mutated for functional analysis of SRRP53608-BR. Single mutants were created by substituting the residues in red (K377 or R512) with alanine. Residues in blue from F411 to T422 in the lower loop were deleted to generate the ΔF411–T422 deletion mutant.