Significance
Protein functions are often evolved from recombination of a limited suite of protein folds. Engineering desired function into proteins via manipulation of the genes, mimicking natural evolutionary processes, represents a promising approach to synthesizing useful molecular tools. Allosteric proteins play important roles in many biological processes, often as regulatory switches or metabolic checkpoints. In this study, analysis of two natural allosteric proteins from different organisms revealed comparable yet distinct regulatory mechanisms via conformation rearrangement. Interchanging gene segments encoding for the nonhomologous regulatory elements of the two proteins results in new functional proteins with switched regulatory behavior. This modular approach represents a step toward a general scheme for construction of functional changes in proteins.
Keywords: protein engineering, allostery, bifunctional enzyme, shikimate, synthetic biology
Abstract
Most proteins comprise two or more domains from a limited suite of protein families. These domains are often rearranged in various combinations through gene fusion events to evolve new protein functions, including the acquisition of protein allostery through the incorporation of regulatory domains. The enzyme 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (DAH7PS) is the first enzyme of aromatic amino acid biosynthesis and displays a diverse range of allosteric mechanisms. DAH7PSs adopt a common architecture with a shared (β/α)8 catalytic domain which can be attached to an ACT-like or a chorismate mutase regulatory domain that operates via distinct mechanisms. These respective domains confer allosteric regulation by controlling DAH7PS function in response to ligand Tyr or prephenate. Starting with contemporary DAH7PS proteins, two protein chimeras were created, with interchanged regulatory domains. Both engineered proteins were catalytically active and delivered new functional allostery with switched ligand specificity and allosteric mechanisms delivered by their nonhomologous regulatory domains. This interchangeability of protein domains represents an efficient method not only to engineer allostery in multidomain proteins but to create a new bifunctional enzyme.
Protein allostery is central to the regulation of many biological processes, including ligand transport and metabolic function and control. Allostery occurs when ligand binding at one site communicates with a remote functional site, resulting in a change in protein function. Allosteric regulation of protein functions often involves a complex network of interactions to deliver signals between distal sites. Signal communication is achieved via diverse mechanisms ranging from large conformational changes to subtle changes in protein dynamics (1–3). The understanding of these remote communications is of great interest, particularly in the fields of drug design and protein engineering (4, 5).
Most proteins contain two or more domains (6). These domains and their interactions govern the function of a protein and are considered evolutionary units for modular assembly of new protein architectures (7–9). Biological data suggest that only a limited number of protein folds exist in nature, and protein functions evolve from mutation, duplication, and recombination of ancestral genes under selective pressure (10). Domain recombination via gene fusion events represents one of the major pathways for the evolution of allostery (11–14). This principle has been recognized in many natural systems (12).
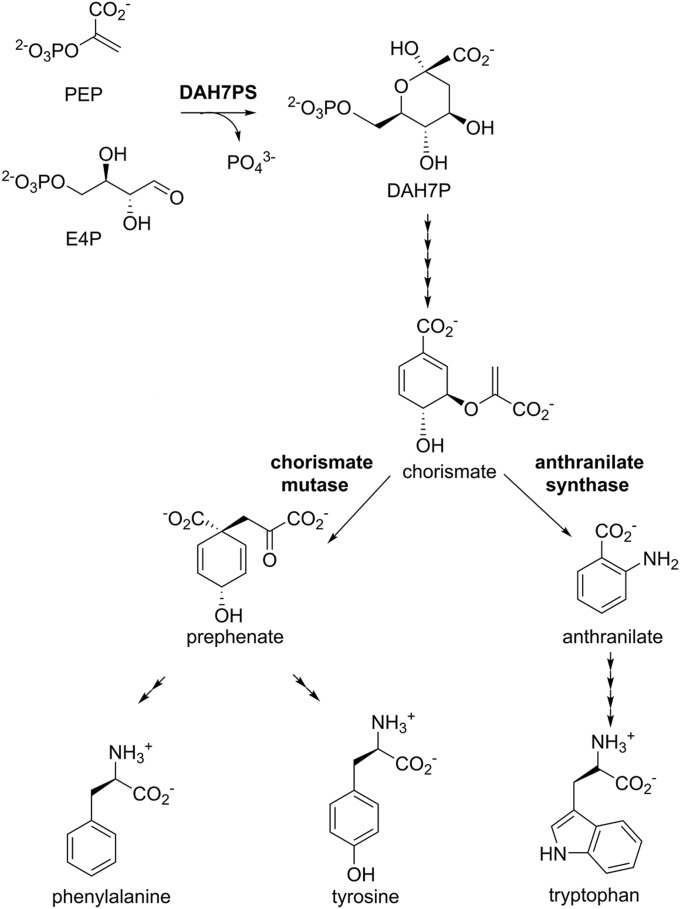
The enzyme 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (DAH7PS) is the first enzyme of the shikimate pathway for the biosynthesis of aromatic compounds. DAH7PS catalyzes the divalent metal-dependent condensation of two carbohydrate precursors, phosphoenolpyruvate (PEP) and d-erythrose 4-phosphate (E4P), to produce DAH7P and phosphate. This enzyme is essential for most microorganisms because of its key role in the biosynthesis of the intermediate chorismate from which the pathway branches to allow formation of prephenate by chorismate mutase (CM) as well as a series of other important metabolites, including aromatic amino acids Phe, Tyr, and Trp (Fig. 1) (15). DAH7PS, situated at the first committed step of the pathway, is often precisely feedback regulated to control pathway flux in response to metabolic demand via a variety of allosteric strategies in different organisms. This allostery ranges from physical gating mechanisms undertaking large conformational changes to highly intricate through-protein dynamic networks (16). The levels of chorismate, prephenate, and the aromatic amino acids are important check points for feedback regulation of the DAH7PS function (16).
Fig. 1.
The shikimate pathway leads to biosynthesis of aromatic amino acids Trp, Phe, and Tyr. DAH7PS catalyzes the first reaction. The pathway branches at chorismate, where CM catalyzes the conversion of chorismate to prephenate en route to formation of Phe and Tyr. The number of arrowheads represents the number of reactions.
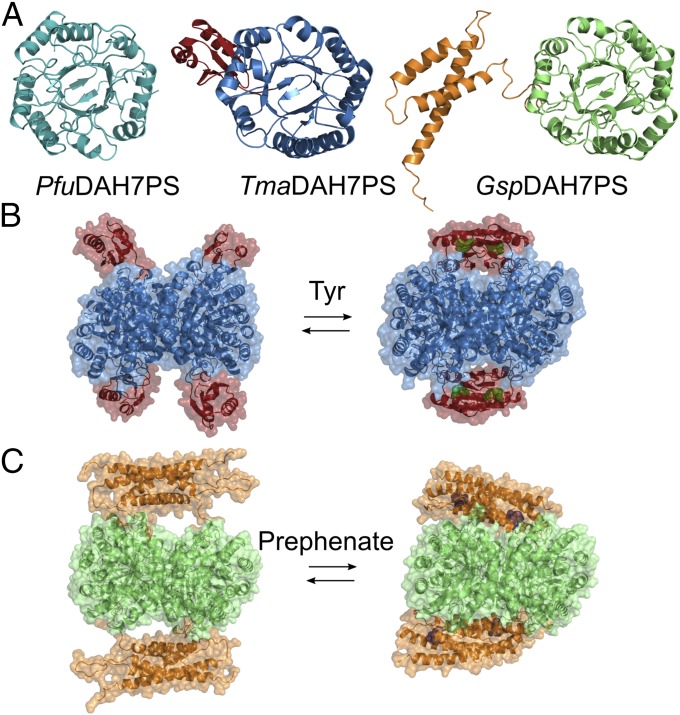
Most DAH7PS enzymes are tetrameric in solution, with each chain comprising a (β/α)8 catalytic barrel. This barrel is frequently decorated with additional structural elements, which are responsible for conferring allostery (16). One major class of DAH7PS enzymes (type Iβ) share a catalytic domain of a structurally uninterrupted classic TIM barrel (β/α)8 fold, and are either unregulated or allosterically regulated (Fig. 2A). The simplest form is composed solely of the barrel without any additional domain, and hence is unregulated. This form of DAH7PS enzymes has been characterized from Pyrococcus furiosus and Aeropyrum pernix (17–19). The regulated DAH7PSs in this group display discrete domains appended to the catalytic barrel at the N or C terminus. These domains are either an ACT-like domain (20) or a CM (AroQ) domain, which possesses CM enzymatic activity. The attached ACT-like and CM domains deliver allostery on a tetrameric DAH7PS scaffold via physical gating of the active site associated with the binding of an allosteric ligand (Fig. 2 B and C) (21–23). This is best exemplified in the well-characterized DAH7PS from Thermotoga maritima (Fig. 2B), which undergoes a remarkable conformational change in response to the presence of Tyr, in which the ACT-like domains from opposing chains dimerize to form a binding site for the allosteric ligand and block substrate access to the active site (21). This enzyme is also inhibited by Phe, although to a reduced extent. Similarly, prephenate binding to the CM domain in DAH7PS from Geobacillus sp. (strain Y412MC61) is associated with the more intimate association between the CM and DAH7PS domains to form a more compact structure and limit catalysis (Fig. 2C) (23).
Fig. 2.
Structure and allostery of DAH7PS. (A) Monomeric units of DAH7PS from P. furiosus (Left), T. maritima (Center), and Geobacillus sp. (Right) share an uninterrupted catalytic barrel. ACT domain in TmaDAH7PS is shown in red; CM domain in GspDAH7PS is shown in orange. Both TmaDAH7PS (B) and GspDAH7PS (C) display significant conformational changes upon ligand binding to the regulatory domain (Tyr is shown as green spheres, and prephenate is shown as purple spheres).
Previously, we have demonstrated the transfer of the allosteric domain of the T. maritima DAH7PS onto the unadorned, unregulated DAH7PS of P. furiosus to confer an allosteric response in the latter DAH7PS (24). We propose that the recruitment of a regulatory domain is a general strategy that, by itself, is sufficient for providing allosteric control of enzymes. To validate this hypothesis and to demonstrate that the key information associated with delivery of allostery resides in the regulatory domain, we explore here the interchangeability of the two distinct regulatory strategies employed by different allosterically controlled DAH7PS enzymes through construction of protein variants that mix and match catalytic and regulatory domains of DAH7PSs from T. maritima and Geobacillus sp. These studies illustrate the remarkable ease with which functional allostery can be acquired by gene fusion events and provide insight into the evolution of modular allostery, in which existing ligand-binding domains or enzymes can be repurposed to provide allostery.
Results
Altered Regulatory and Catalytic Domain Combinations Deliver Functional Proteins.
The wild-type parent proteins TmaDAH7PS and GspDAH7PS share similar homotetrameric quaternary structures and tertiary structures, with each chain composed of an N-terminal regulatory domain (respectively, ACT and CM) attached to a catalytic (β/α)8 barrel housing the active site. The TmaDAH7PS and GspDAH7PS catalytic barrels share a moderate sequence identity of 56% (Fig. S1). Two protein variants were created with exchanged regulatory and catalytic domains based on the analysis of wild-type sequences and architectures. Both these domain-swapped variants delivered DAH7PS catalysis, albeit with some alterations in their catalytic efficiencies (Table 1). Compared with the wild-type TmaDAH7PS, GspCM-TmaDAH7PS, which shares the same catalytic core, exhibited impaired activity with significantly decreased kcat/KM values for both PEP and E4P substrates, implying there may be some restriction of access for substrates to the catalytic center introduced by the fused CM domains. The origin of this may lie in the fact that the CM domain is dimeric both in the absence and presence of the allosteric ligand, whereas the ACT-like domain only dimerizes upon ligand binding (21, 23). Hence, perhaps unsurprisingly, the adoption of the more restricted GspCM domain to the TmaDAH7PS catalytic core is accompanied by attenuation of catalysis, whereas TmaACT-GspDAH7PS displayed an approximately twofold boost in catalytic efficiency compared with the wild-type GspDAH7PS. That physical constraints restrict catalysis by the DAH7PS core barrel is supported by the higher activity displayed by truncated forms of TmaDAH7PS and GspDAH7PS in which, respectively, the ACT and CM domains have been removed (21, 23).
Table 1.
Kinetics parameters for the chimeras and parent proteins
| DAH7PS activity | CM activity | |||||||
| Protein | KMPEP, µM | KME4P, µM | kcat, s−1 | kcat/KMPEP, µM−1s−1 | kcat/KME4P, µM−1s−1 | KMchorismate, µM | kcat, s−1 | kcat/KM, µM−1s−1 |
| GspCM-TmaDAH7PS | 34 ± 2.7 | 19 ± 1.8 | 4.8 ± 0.1 | 0.14 | 0.25 | 190 ± 16 | 4.1 ± 0.1 | 0.02 |
| TmaACT-GspDAH7PS | 57 ± 1.6 | 54 ± 1.4 | 60 ± 2 | 1.05 | 1.11 | NA | NA | NA |
| GspDAH7PS | 45 ± 4 | 61 ± 8 | 28 ± 0.9 | 0.62 | 0.46 | 98 ± 7 | 3.4 ± 0.1 | 0.03 |
| TmaDAH7PS | 8.4 ± 0.7 | 15 ± 1 | 14 ± 0.3 | 1.67 | 0.93 | NA | NA | NA |
NA, not applicable for the specified enzyme activity.
Unlike the ACT domain, which has only a ligand-binding role, the regulatory domain of GspDAH7PS has both catalytic and regulatory functions (23). The CM catalytic activity was comparably maintained when the domain was transferred to the alternative catalytic barrel in GspCM-TmaDAH7PS (Table 1), although this chimera displayed an approximately twofold increase in KM for chorismate compared with wild-type GspDAH7PS. As expected, with no CM domain, the variant TmaACT-GspDAH7PS displayed only DAH7PS activity.
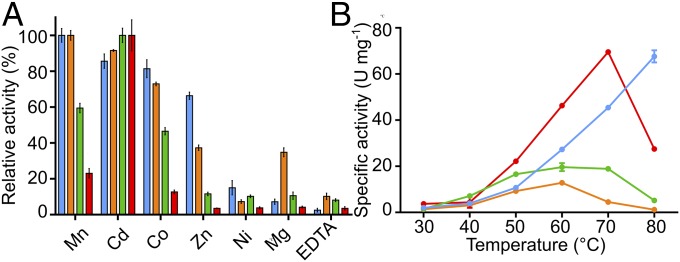
As with all wild-type DAH7PS enzymes characterized to date, the catalytic activities of the chimeras depend on presence of a divalent metal ion (Fig. 3A). Mn2+ is the most activating metal ion for the wild-type TmaDAH7PS, whereas Cd2+ delivers maximal activity for GspDAH7PS (21, 23). A range of metal ions were tested with the protein chimeras. Largely as expected, each of the DAH7PS catalytic cores reflected its inherent metal preference, with TmaACT-GspDAH7PS highly favoring Cd2+, whereas GspCM-TmaDAH7PS showed more than 90% activity in the presence of Mn2+ or Cd2+.
Fig. 3.
Catalytic activity of the parent proteins and chimeras. (A) DAH7PS activities of TmaDAH7PS (blue), GspDAH7PS (green), TmaACT-GspDAH7PS (red), and GspCM-TmaDAH7PS (orange), in the presence metal ions or EDTA. (B) The effect of temperature on specific activity of the four enzymes in the same color coding as in A.
Consistent with the thermophilic properties of the wild-type enzymes, both enzymes became more active at elevated temperatures, reaching their optimal activity at temperatures above 60 °C (Fig. 3B). Intriguingly, activity of TmaACT-GspDAH7PS was enhanced significantly at elevated temperatures compared with the wild-type GspDAH7PS. On the other hand, GspCM-TmaDAH7PS appeared to be least active across all temperatures. Proteins with the GspCM domain, chimeric and wild-type, generally tend to display lower activities than proteins with the TmaACT domain at the temperatures tested. This change in activity profile may relate to the structural difference in the regulatory domains and the different inherent optimal temperatures of the parent proteins.
Allosteric Inhibitor Preference Resides in the Regulatory Domain.
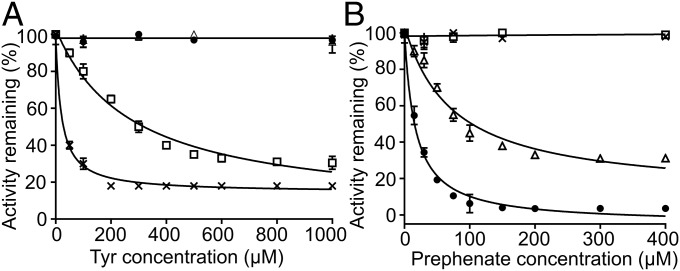
To test the effect of potential allosteric ligands, inhibition assays were performed for both DAH7PS chimeras and compared with the response of the wild-type proteins (Fig. 4 and Fig. S2). Consistent with the presence of a regulatory domain in both chimeras, inhibition of both enzymes was observed. Presenting the ACT domain, the DAH7PS activity of TmaACT-GspDAH7PS was reduced by 70% at high Tyr concentrations and by ∼30% at high Phe concentrations. This difference in ligand sensitivity for Tyr and Phe is comparable with that of the wild-type TmaDAH7PS, for which Tyr exhibits more significant inhibitory effect than Phe, although the IC50 value of Tyr for the chimera (210 µM) is almost 10-fold higher than that of the wild-type protein (22.5 µM). The presence of the CM domain in GspCM-TmaDAH7PS delivered sensitivity toward prephenate, although the inhibition was less profound than that for the wild-type GspDAH7PS. The presence of 77 µM prephenate reduced DAH7PS activity to 50% of its uninhibited value, compared with 20 µM required for the same attenuation of wild-type protein catalytic activity. The maximum level of inhibition was also altered; the chimeric protein retained 31% activity at higher prephenate concentrations in comparison with the 4% residual activity observed for the wild-type enzyme.
Fig. 4.
Inhibition of DAH7PS activity for the parent proteins and chimeras. (A) TmaDAH7PS (×) and TmaACT-GspDAH7PS (□) displayed sensitivity toward Tyr. (B) GspDAH7PS (●) and GspCM-TmaDAH7PS (△) displayed impaired catalysis with addition of prephenate.
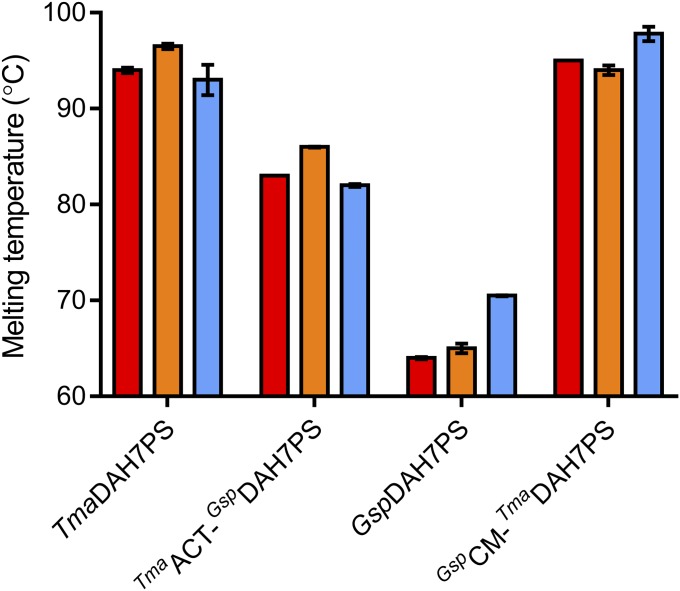
Binding of allosteric ligands stabilizes the parent proteins and the chimeras (Fig. 5 and Fig. S3). In the presence of inhibitors, the melting temperatures of both chimeras increased by around 3 °C, in close agreement with the degree of stabilization for the wild-type proteins. Interestingly, the thermostability of TmaACT-GspDAH7PS improved almost 20 °C compared with the wild-type GspDAH7PS. On the other hand, the stability of the GspCM-TmaDAH7PS appeared to be largely determined by the TmaDAH7PS core, with the denaturing event occurring at above 96 °C. The thermostability profiles exhibited by the chimeras largely reflect the difference in inherent thermostability of the wild-type proteins, remarkably, even when the transferred domain is only a fraction (approximately one-third) of the core DAH7PS barrel, as in TmaACT-GspDAH7PS.
Fig. 5.
Thermostability of the parent proteins and chimeras in the absence (red) and presence of Tyr (orange) or prephenate (blue).
The Structural Changes of Allostery Are Transferred with the Regulatory Domains.
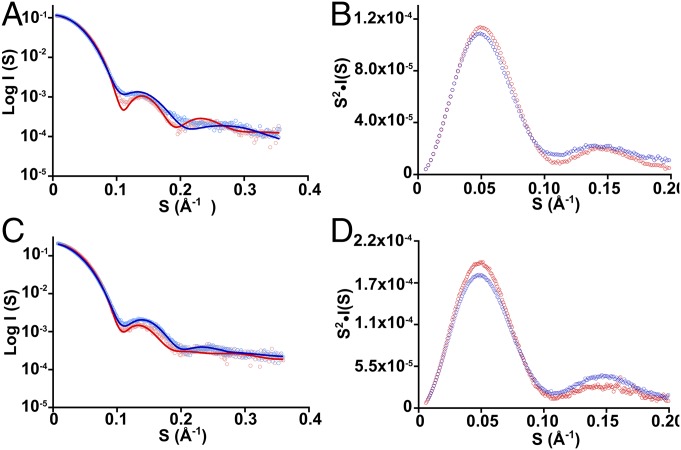
The allosteric mechanisms of both parental proteins involve conformational rearrangements as observed in solution by small-angle X-ray scattering (SAXS) experiments (21, 23). To assess the allosteric mechanisms employed by the chimeras, SAXS experiments were performed. In the presence of Tyr, TmaACT-GspDAH7PS became more compact, with a reduced Rg value (derived from Guinier plot) of 32.8 ± 0.2 Å compared with the Rg of the apo form 34.0 ± 0.2 Å (Fig. 6 and Table S2). The Kratky plot displayed a more defined curve in the presence of Tyr than without Tyr, consistent with the decreased flexibility of the protein. The GspCM-TmaDAH7PS also showed a conformational change with addition of prephenate, with Rg decreasing from 36.9 ± 0.2 Å to 34.8 ± 0.2 Å, consistent also with the more defined Kratky plot.
Fig. 6.
SAXS profiles of the chimeras. (A) SAXS profiles of TmaACT-GspDAH7PS were measured in the absence (light blue circles) and presence (pink circles) of Tyr. Theoretical scatterings were calculated from Tyr-free (blue line) and Tyr-bound (red line) TmaDAH7PS crystal structures. (B) Kratky plot of TmaACT-GspDAH7PS SAXS profiles in the absence (blue circles) and presence (red circles) of Tyr. (C) SAXS profiles of GspCM-TmaDAH7PS were measured in the absence (light blue circles) and presence (pink circles) of prephenate. Theoretical scatterings were calculated from prephenate-free model (blue line) and prephenate-bound (red line) crystal structure of GspDAH7PS. (D) Kratky plot of GspCM-TmaDAH7PS SAXS profiles in the absence (blue circles) and presence (red circles) of prephenate. I, scattering intensity; S, magnitude of scattering vector (where S = 4πsinθ/λ, 2θ is the scattering angle and λ is the wavelength of the radiation).
To compare these conformational changes observed in the protein chimeras with those observed for the wild-type proteins, the SAXS profiles were fitted with the calculated theoretical scattering from crystal structures or homology model of each parent protein (Fig. 6 and Table S2). In the absence of Tyr, scattering profiles of TmaACT-GspDAH7PS presented a good fit with the open form of the TmaDAH7PS crystal structure, but not the closed form. Conversely, in the presence of Tyr, the scattering profile of TmaACT-GspDAH7PS gave a better fit with the closed, Tyr-bound form of TmaDAH7PS than with the open, apo form. In the absence of prephenate, the scattering profile of GspCM-TmaDAH7PS fits closely with the open-form model of the wild-type GspDAH7PS and poorly with the closed, prephenate-bound form of GspDAH7PS. In the presence of prephenate, GspCM-TmaDAH7PS is better fit by the closed, prephenate-bound form of GspDAH7PS than by the open, apo form.
Discussion
Design of proteins with new properties and functions is an important goal of biotechnology. The modular feature of many natural proteins suggests that common building domains and modules are likely to have the evolutionary advantages of being autonomous and portable, offering simplicity in recombination to generate new functions by allowing transfer of information through their interactions (25). Statistical coupling analysis revealed that the recombination of protein domains relies on the networks of coevolving amino acids involved in the allosteric communication and that these networks display strong connectivity, proposing the feasibility of engineering artificial allosteric systems by transfer of allosteric networks between proteins (5, 26). Although this transferability has not been extensively explored, a few studies indicate its plausibility. For example, a modulating maltose-binding protein was fused to an unrelated enzyme β-lactamase, introducing maltose regulation to the unregulated β-lactamase (27–29). In another example, ultrasensitive molecular switches were built by modular recombination of multiple SH3–peptide autoinhibitory interactions on WASP to introduce strong cooperativity with respect to the ligand SH3 (30, 31). Also, an Escherichia coli dihydrofolate reductase (DHFR) was coupled with a light-sensing protein from plants to generate a light-sensitive DHFR (5). These studies demonstrate that methods of mix and match between modular components can be effective ways to create substantial functional changes associated with allostery and to expand the repertoire of artificial proteins.
What we demonstrate here is the interchangeability of regulatory domains in a homooligomeric protein. By simple gene recombination of contemporary sequences, we can interchange the Tyr-binding ACT domain on one DAH7PS with a structurally nonhomologous prephenate-binding CM domain from another DAH7PS. This interchange functionally swaps allosteric regulation in both DAH7PS enzymes elicited by formation of, or changes to, a dimeric structure upon binding of the appropriate allosteric effector. What is somewhat surprising is that gene fusion to functionally link these contemporary domains requires no modifications to the core catalytic barrels. Fusion of the regulatory domain nevertheless inflicts some catalytic penalty on the barrel, and the degree of compensated activity may be associated with the nature of the extended domains. We note that the functional domain-swapped chimeras developed here are quite distinct from other examples where chimeras have been generated by the modification of discrete structural elements on structurally homologous domains to alter allosteric ligand specificity (32–35). We have interchanged two structurally and functionally nonhomologous regulatory modules of contemporary DAH7PS scaffolds. That fully functional enzymes are generated highlights the conservation of allosteric strategy in these contemporary proteins, which is delivered by structurally diverse solutions (or gene fusions) (21, 23).
Despite the ease of functional allostery interchange, there are enabling features of the contemporary DAH7PS scaffold that are important in accommodating the recombination and for maintaining allosteric networks. Core barrel oligomerization is a prerequisite for allostery that involves dimerization of the regulatory module (17, 21, 23, 36). The DAH7PS barrel homotetramer adopts an overall conformation that supports the allosteric function, with diagonally opposite chains delivering the regulatory domain on either side of the tetramer plane (Fig. 2). This tetrameric assembly is a feature of type Iβ DAH7PS, which is shared by both regulated and unregulated DAH7PSs (19, 21, 23, 37) and a related enzyme, 3-deoxy-d-manno-2-octulosonate 8-phosphate synthase (38). Intriguingly, quaternary structure is delicately balanced in this DAH7PS enzyme class. Removal of the ACT-like domain from TmaDAH7PS results in dimerization and the (unregulated) PfuDAH7PS is rendered dimeric by a single amino acid substitution (19, 21). Association of dimers into tetramers was probably followed by acquisition of terminal regulatory domains (17). Thus, it appears that the functional gene fusion to deliver allostery was facilitated serendipitously by the adoption of the appropriate homotetrameric catalytic template.
The barrels are not highly conserved overall, but there do appear to be some sequence elements that are associated with delivering allostery in both systems that are shared between both GspDAH7PS and TmaDAH7PS. The key hydrogen-bond contacts between either the ACT domain or the CM domain with the catalytic DAH7PS domain reside mostly at the C terminus of the diagonally adjacent barrel (Fig. S4). In the ligand-bound form of TmaDAH7PS, major hydrogen bonds are formed between an ACT domain and the barrel from the adjacent chain, including Asp51 and Asp309, Ser55 and Arg277, and Asp57 and Glu304. Due to the asymmetrical nature of the domain arrangement across the vertical plane in the crystal structure of GspDAH7PS, each CM polypeptide chain communicates with its adjacent barrel through different hydrogen-bonding interactions. Key interactions are found between the α1-α2 loop of the CM domain and the C terminus of the barrel, including Arg52 and Asp326, as well as Tyr46 and Glu29 on one end of the barrel, plus Gly42 and Arg157 on the other end.
Sequence alignment between TmaDAH7PS and GspDAH7PS suggests that these key regions on the catalytic barrel involved in the communication of the allosteric information are highly similar (Fig. S1). Indeed, these regions are generally well conserved across the type Iβ family (Fig. S4C), which is consistent with the involvement of these contact regions in DAH7PS catalysis. All contact regions are located on the top side of the barrel close to the active site. Key residues of the β2-α2 and β8-α8 loops form direct hydrogen-bonding interactions with the substrates PEP and E4P, and segments of loop β1-α1 and β7-α7 appear to secure the correct positioning of those residues that interact with substrates. Together, the DAH7PS barrel appears to be a versatile platform for additional functions, which tend to be developed conveniently by hijacking the conserved features of the active site so that allostery can be created with ease when interdomain interactions are reestablished in the presence of ligand. Additionally, some conservation of residue character for the N-terminal regulatory domains (charged, polar, and nonpolar) may also contribute to the establishment of an allosteric network between DAH7PS and regulatory domain, despite very different secondary and tertiary structures of the ACT and CM domains. The linkers between the DAH7PS and its regulatory domain share a remarkably high degree of conservation, possibly because of the conformational restrictions for the hinging motions occurring on binding the allosteric effectors. This conservation in the flexible linker region may be indicative of its functional importance. Protein stability may have also contributed to the success of domain swapping, as proteins with enhanced stability are believed to have greater capacity for modifications (39). These underlying features and modular scaffold of the DAH7PS family appear to support the acquisition of functional variations.
In addition to the transfer of allostery, enzymatic activity of the CM domain was also transferred into the GspCM-TmaDAH7PS chimera. Remarkably, thermostability was determined by the more thermophilic component of the chimera, even when that component, as in the case of the ACT domain of the DAH7PS from the hyperthermophilic T. maritima, was transferred onto the much larger DAH7PS domain from the less thermophilic Geobacillus sp.—the proverbial tail wagging the dog. The generality of this phenomenon as a mechanism for inducing thermostability into more mesophilic enzymes remains to be elucidated.
From one enzyme and two distinct regulatory domains we have four allosterically regulated enzymes, two occurring in nature and two created in the laboratory, one of which is additionally a bifunctional enzyme. The engineering of allosteric control, enzymatic activities, and thermostability by domain swapping illustrates that homologous (β/α)8 structures can tolerate new combinations with structurally and functionally nonhomologous regulatory domains. Future investigations into the details of the corresponding allosteric network experimentally and computationally are important for the design and optimization of allosteric systems with high performance.
Materials and Methods
Design and Preparation of Protein Variants.
Crystal structures of the TmaDAH7PS and GspDAH7PS suggest similar architecture and arrangement of the homotetramers, with the catalytic barrels connected to their respective N-terminal regulatory domains via a β-hairpin and a flexible linker region. The linker region is crucial for the interaction between regulatory and catalytic domains in both enzymes and to allow appropriate conformational change between active and inhibited states (21). The amino acid regions that incorporate regulatory domains, linker regions, and catalytic domains of TmaDAH7PS and GspDAH7PS are identified individually from sequence and structure alignments (Fig. S1). Two protein variants with exchanged regulatory and catalytic domains were designed: TmaACT-GspDAH7PS, with the regulatory domain and linker region from TmaDAH7PS (residues 1 to 93) and the catalytic domain from GspDAH7PS (residues 118 to 362); and the complementary chimera GspCM-TmaDAH7PS, with the regulatory domain and linker region from GspDAH7PS (residues 1 to 117) and the catalytic domain from TmaDAH7PS (residues 94 to 338). Both constructs were created by amplifying each segment from the corresponding parent wild-type gene with overlap and by fusing the amplified products (Table S1). The fused gene fragments encoding each chimeric protein were cloned using Gateway® technique. TmaACT-GspDAH7PS was transformed in pDEST14 and expressed in BL21(DE3)* cells, and its expression and purification were performed following previously published procedures for TmaDAH7PS (21). GspCM-TmaDAH7PS was transformed in pDEST15 and expressed in BL21(DE3)pLysS cells using the same expression conditions, and purified using an GSTrap HP column following published procedures for the truncated GspDAH7PS (23).
Enzyme Kinetics.
Kinetics parameters for DAH7PS and/or CM activities of the chimeric proteins were determined following previously described procedures by measuring consumption of chorismate at 274 nm or PEP at 232 nm, with each reaction containing 0.05 µM enzyme (23). For DAH7PS assays, the concentration of E4P was varied between 5 µM and 400 µM while PEP was held at 295 µM, and the concentration of PEP was varied between 6 µM and 350 µM while E4P was fixed at 310 µM. Reactions contained enzyme, PEP, and 100 µM Mn2+ (for proteins containing the catalytic barrel from T. maritima) or Cd2+ (for proteins containing the catalytic barrel from Geobacillus sp.), and were equilibrated in 50 mM 1,3-bis[tris(hydroxymethyl)methylamino] propane (BTP) buffer at 60 °C (pH 7.4) before E4P was added to initiate the reaction. For CM assays, 9 to 400 µM chorismate was used to initiate the reaction in the same BTP buffer at 50 °C. Metal dependency studies were performed using the same DAH7PS assay described above in the presence of saturating PEP and E4P, with a range of metal ions or EDTA at 100 µM. For inhibition studies, assay solutions contained 283 µM PEP, 308 µM E4P, 100 µM Mn2+ or Cd2+, and 0 to 1 mM Tyr/Phe or 0 to 400 µL prephenate in 50 mM BTP (pH 7.4 at 60 °C).
Thermal Activity and Stability Measurements.
Activity profiles of the chimeric proteins and parent proteins at elevated temperatures (30 to 80 °C) were assessed using the same methods as for kinetics measurements of DAH7PS activity. Each reaction contained 0.05 µM enzyme, 100 µM appropriate metal ion, 215 µM PEP, and 227 µM E4P in 50 mM BTP buffer. All buffers were at pH 7.4 at the temperatures tested. Enzyme activities are specified in U (1 U = consumption of 1 μmol substrate per minute). Specific enzyme activities are given as U·mg−1. Melting temperatures were determined by using differential scanning fluorimetry with an iCycler iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad). Protein samples (0.1 mg·mL−1) were prepared in storage size-exclusion chromatography buffer with SYPRO Orange dye in the absence or presence of ligands (1 mM Tyr/Phe or 500 μM prephenate). Controls were similarly prepared for each condition, with buffer in place of protein. The temperature was set to increase from 20 to 98 °C at a rate of 1 °C/min, and fluorescence was measured in 0.2 °C increments.
SAXS.
SAXS measurements were collected at the Australian Synchrotron SAXS/wide-angle X-ray scattering (WAXS) beamline using the setup previously described (21, 23). Scattering data were collected at 25 °C following elution of the protein samples (9 mg/mL) from a size-exclusion chromatography column (Superdex 200_Increase 5/150), which was preequilibrated with buffer containing 10 mM BTP (pH 7.4), 100 mM KCl, and 200 µM PEP, in the absence or presence of inhibitors (1 mM Tyr or 500 µM prephenate). Raw data were processed as described for wild-type proteins using Scatterbrain (21). Scattering intensity versus magnitude of scattering vector of each protein was generated with Primus, and plots from apo- and liganded proteins were scaled (40). Theoretical scattering profiles were generated from published crystal structures of wild-type TmaDAH7PS 1RZM (apo) and 3PG9 (Tyr), homology model of wild-type GspDAH7PS (apo), and crystal structure 5J6F (prephenate) (21, 23, 37). They were fitted with corresponding experimental scatterings using CRYSOL (41).
Sequence Analysis.
Sequence alignments of the wild-type PfuDAH7PS, TmaDAH7PS, and GspDAH7PS were generated with Clustal Omega (42) and formatted with ESPript 3 (43). A total of 2,523 sequences from the DAH7PS family of interest (type Iβ) were obtained from Pfam (PF00793) (44) and aligned using SeaView 4 (45), and conserved residues were identified and graphed with WebLogo3 (46).
Supplementary Material
Acknowledgments
Y.F. is a recipient of a University of Canterbury Doctoral Scholarship. This work was supported by grants from the Marsden Fund (UOC1105), New Zealand Synchrotron Group Ltd., and the SAXS/WAXS beamline at the Australian Synchrotron.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717621115/-/DCSupplemental.
References
- 1.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(6761, Suppl):C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 2.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 4.Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr Opin Struct Biol. 2006;16:102–108. doi: 10.1016/j.sbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Lee J, et al. Surface sites for engineering allosteric control in proteins. Science. 2008;322:438–442. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma B, Tsai CJ, Haliloğlu T, Nussinov R. Dynamic allostery: Linkers are not merely flexible. Structure. 2011;19:907–917. doi: 10.1016/j.str.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doolittle RF. The multiplicity of domains in proteins. Annu Rev Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- 8.Patthy L. Modular assembly of genes and the evolution of new functions. Genetica. 2003;118:217–231. [PubMed] [Google Scholar]
- 9.Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol. 2004;14:208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- 11.Peracchi A, Mozzarelli A. Exploring and exploiting allostery: Models, evolution, and drug targeting. Biochim Biophys Acta. 2011;1814:922–933. doi: 10.1016/j.bbapap.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Bashton M, Chothia C. The generation of new protein functions by the combination of domains. Structure. 2007;15:85–99. doi: 10.1016/j.str.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Ostermeier M, Benkovic SJ. Evolution of protein function by domain swapping. Adv Protein Chem. 2000;55:29–77. doi: 10.1016/s0065-3233(01)55002-0. [DOI] [PubMed] [Google Scholar]
- 14.Pasek S, Risler JL, Brézellec P. Gene fusion/fission is a major contributor to evolution of multi-domain bacterial proteins. Bioinformatics. 2006;22:1418–1423. doi: 10.1093/bioinformatics/btl135. [DOI] [PubMed] [Google Scholar]
- 15.Bentley R. The shikimate pathway–A metabolic tree with many branches. Crit Rev Biochem Mol Biol. 1990;25:307–384. doi: 10.3109/10409239009090615. [DOI] [PubMed] [Google Scholar]
- 16.Light SH, Anderson WF. The diversity of allosteric controls at the gateway to aromatic amino acid biosynthesis. Protein Sci. 2013;22:395–404. doi: 10.1002/pro.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schofield LR, et al. Substrate ambiguity and crystal structure of Pyrococcus furiosus 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase: An ancestral 3-deoxyald-2-ulosonate-phosphate synthase? Biochemistry. 2005;44:11950–11962. doi: 10.1021/bi050577z. [DOI] [PubMed] [Google Scholar]
- 18.Zhou L, et al. Structure and characterization of the 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Aeropyrum pernix. Bioorg Chem. 2012;40:79–86. doi: 10.1016/j.bioorg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nazmi AR, Schofield LR, Dobson RCJ, Jameson GB, Parker EJ. Destabilization of the homotetrameric assembly of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from the hyperthermophile Pyrococcus furiosus enhances enzymatic activity. J Mol Biol. 2014;426:656–673. doi: 10.1016/j.jmb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Lang EJ, Cross PJ, Mittelstädt G, Jameson GB, Parker EJ. Allosteric ACTion: The varied ACT domains regulating enzymes of amino-acid metabolism. Curr Opin Struct Biol. 2014;29:102–111. doi: 10.1016/j.sbi.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Cross PJ, Dobson RC, Patchett ML, Parker EJ. Tyrosine latching of a regulatory gate affords allosteric control of aromatic amino acid biosynthesis. J Biol Chem. 2011;286:10216–10224. doi: 10.1074/jbc.M110.209924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Light SH, Halavaty AS, Minasov G, Shuvalova L, Anderson WF. Structural analysis of a 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase with an N-terminal chorismate mutase-like regulatory domain. Protein Sci. 2012;21:887–895. doi: 10.1002/pro.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nazmi AR, et al. Interdomain conformational changes provide allosteric regulation en route to chorismate. J Biol Chem. 2016;291:21836–21847. doi: 10.1074/jbc.M116.741637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cross PJ, Allison TM, Dobson RCJ, Jameson GB, Parker EJ. Engineering allosteric control to an unregulated enzyme by transfer of a regulatory domain. Proc Natl Acad Sci USA. 2013;110:2111–2116. doi: 10.1073/pnas.1217923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 26.Hatley ME, Lockless SW, Gibson SK, Gilman AG, Ranganathan R. Allosteric determinants in guanine nucleotide-binding proteins. Proc Natl Acad Sci USA. 2003;100:14445–14450. doi: 10.1073/pnas.1835919100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostermeier M. Engineering allosteric protein switches by domain insertion. Protein Eng Des Sel. 2005;18:359–364. doi: 10.1093/protein/gzi048. [DOI] [PubMed] [Google Scholar]
- 28.Betton J-M, Jacob JP, Hofnung M, Broome-Smith JK. Creating a bifunctional protein by insertion of β-lactamase into the maltodextrin-binding protein. Nat Biotechnol. 1997;15:1276–1279. doi: 10.1038/nbt1197-1276. [DOI] [PubMed] [Google Scholar]
- 29.Guntas G, Ostermeier M. Creation of an allosteric enzyme by domain insertion. J Mol Biol. 2004;336:263–273. doi: 10.1016/j.jmb.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 30.Dueber JE, Mirsky EA, Lim WA. Engineering synthetic signaling proteins with ultrasensitive input/output control. Nat Biotechnol. 2007;25:660–662. doi: 10.1038/nbt1308. [DOI] [PubMed] [Google Scholar]
- 31.Dueber JE, Yeh BJ, Chak K, Lim WA. Reprogramming control of an allosteric signaling switch through modular recombination. Science. 2003;301:1904–1908. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- 32.Cunin R, Rani CS, Van Vliet F, Wild JR, Wales M. Intramolecular signal transmission in enterobacterial aspartate transcarbamylases II. Engineering co-operativity and allosteric regulation in the aspartate transcarbamylase of Erwinia herbicola. J Mol Biol. 1999;294:1401–1411. doi: 10.1006/jmbi.1999.3316. [DOI] [PubMed] [Google Scholar]
- 33.Pawlyk AC, Pettigrew DW. Transplanting allosteric control of enzyme activity by protein-protein interactions: Coupling a regulatory site to the conserved catalytic core. Proc Natl Acad Sci USA. 2002;99:11115–11120. doi: 10.1073/pnas.132393599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Page MJ, Carrell CJ, Di Cera E. Engineering protein allostery: 1.05 A resolution structure and enzymatic properties of a Na+-activated trypsin. J Mol Biol. 2008;378:666–672. doi: 10.1016/j.jmb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helmstaedt K, Heinrich G, Lipscomb WN, Braus GH. Refined molecular hinge between allosteric and catalytic domain determines allosteric regulation and stability of fungal chorismate mutase. Proc Natl Acad Sci USA. 2002;99:6631–6636. doi: 10.1073/pnas.092130899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartmann M, et al. Evolution of feedback-inhibited beta/alpha barrel isoenzymes by gene duplication and a single mutation. Proc Natl Acad Sci USA. 2003;100:862–867. doi: 10.1073/pnas.0337566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shumilin IA, Bauerle R, Wu J, Woodard RW, Kretsinger RH. Crystal structure of the reaction complex of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Thermotoga maritima refines the catalytic mechanism and indicates a new mechanism of allosteric regulation. J Mol Biol. 2004;341:455–466. doi: 10.1016/j.jmb.2004.05.077. [DOI] [PubMed] [Google Scholar]
- 38.Allison TM, et al. An extended β7α7 substrate-binding loop is essential for efficient catalysis by 3-deoxy-D-manno-octulosonate 8-phosphate synthase. Biochemistry. 2011;50:9318–9327. doi: 10.1021/bi201231e. [DOI] [PubMed] [Google Scholar]
- 39.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci USA. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 41.Svergun D, Barberato C, Koch MH. CRYSOL–A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 42.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finn RD, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 46.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.