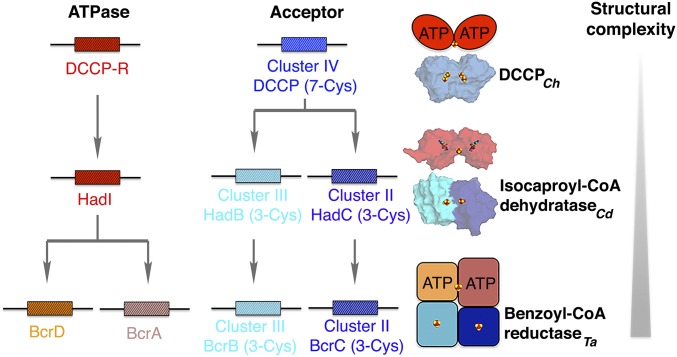
Fig. 5.
Architecture of DCCP homologs. Structural genes are indicated as boxes with distinct colors. Crystal structures of isocaproyl-CoA dehydratase (HadB and HadC; PDB ID code 3O3M) (14) and its activator (HadI; PDB ID code 4EHU) (42) are indicated. Benzoyl-CoA reductase is modeled as suggested in ref. 26. Fe/S-clusters are shown as spheres with iron in red and sulfur in yellow. AMPPNP bound in the dehydratase activator is shown as sticks. Cd: Clostridium difficile; Ta: Thauera aromatica.