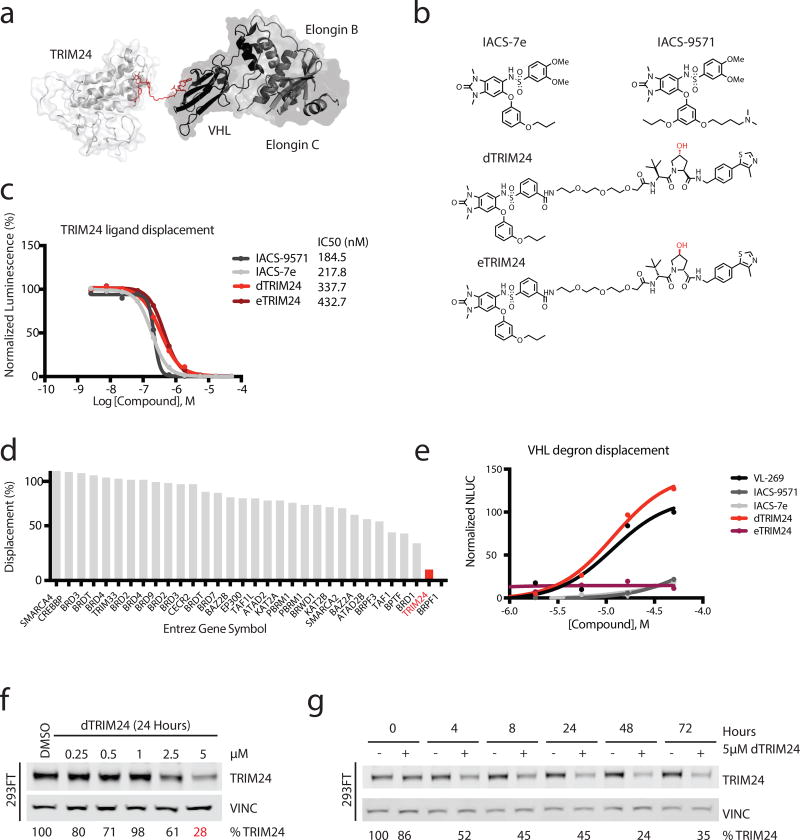
Figure 1. Design and chemical characterization of dTRIM24 as a degrader of TRIM24.
(a) Crystal structure of TRIM24 PHD-BROMO bound by IACS-9571 in proximity to the crystal structure of VHL bound to VL-269 to model a strategy for building the degrader (PDB: 4YC9 and 4W9H). (b) Chemical structures of IACS-7e, IACS-9571, dTRIM24 and eTRIM24 with stereocenter of the VHL ligand component highlighted in red. (c) TRIM24 ligand displacement assay (values represent means normalized to DMSO calculated from n=2 technical replicates). n=3 independent experiments with one representative experiment shown. (d) dTRIM24 in vitro binding assay to panel of human bromodomain proteins by single point screening at 1 µM dTRIM24 in singlicate (BromoScan). (e) Cellular VHL degron displacement assay (values represent means normalized to 50 µM VL-269 calculated from n=2 technical replicates). n=2 independent experiments with one representative experiment shown. (f) Immunoblot of TRIM24 and Vinculin following 24 hours of incubation with the indicated concentrations of dTRIM24 in 293FT cells. (g) Immunoblot of TRIM24 and Vinculin following treatment of 293FT cells with 5 µM dTRIM24 for the indicated incubation times. For f–g, percentages were calculated by normalization of the band intensity to the loading control and relative to DMSO at each timepoint. n=3 independently conducted experiments with one representative experiment shown. Full immunoblots shown in Supplementary Fig. 9.