Abstract
Purpose
To identify the somatic mutated genes for optimal targets of non-small-cell lung cancer after resistance to osimertinib treatment.
Patients and Methods
Study patients all had advanced lung adenocarcinoma and acquired resistance to osimertinib as a second- or third-line treatment. These patients had harboring EGFR T790M mutation before osimertinib treatment, which was confirmed by Amplification Refractory Mutation System (ARMS) PCR or Next-Generation Sequencing (NGS). After resistance to osimertinib treatment, tumor tissue was collected by core needle biopsy. DNA was extracted from 15 × 5 um sliced section of formalin-fixed paraffin-embedded (FFPE) material and NGS was done. The genetic changes were analyzed.
Results
A total of 9 Chinese patients were studied, 5 females and 4 males, age 51–89 years. After progression with osimertinib treatment, core needle biopsy was performed and next-generation sequencing was performed. Nine patients had harboring 62 point mutations, 2 altered gene copies, 2 amplifications, and 1 EML4-ALK gene fusion. No MET or HER2 amplification was found in this cohort study. Nine patients still maintained initial EGFR 19 del or L858R activating mutations, while 7 of them kept EGFR T790M mutations. Among the 7 patients, 5 had secondary EGFR C797S and/or C797G mutations, which all happened in the same allele with T790M mutation. All patients were treated with targets therapies, chemotherapy, or best supportive care (BSC) in accordance with NGS genetic results and patients' performance status; 7 of them are still alive and 2 of them died of disease progression at last follow-up.
Conclusions
EGFR C797S/G mutation and the same one presented on the same allele with EGFR T790M mutation were the most common mutation feature and played a key role in resistance to osimertinib in Chinese patients with NSCLC. Tumor cells losing T790M mutation and maintaining EGFR activating mutation might benefit from first-generation EGFR-TKI treatment.
1. Introduction
Epidermal growth factor receptor (EGFR) T790M mutation is the most common genetic change for patients of non-small-cell lung cancer (NSCLC) harboring EGFR after resistance to first-generation EGFR tyrosine kinase inhibitor (TKI) [1]. The substitution of threonine with methionine at amino acid position 790 (T790M), which reduces the ability of ATP-competitive reversible EGFR-TKI binding to EGFR tyrosine kinase domain, results in cancer cells resistant to gefitinib and erlotinib [1]. Osimertinib (Tagrisso, AZD9291, AstraZeneca) is the only FDA approved drug for lung cancer patients harboring EGFR T790M mutation. After a median of 9.6–11.0 months' remission with osimertinib treatment, tumors will inevitably have progress. Although a lot of studies had been done, the molecular mechanisms of resistance are not yet fully understood [2, 3].
Next-generation sequencing (NGS) is a cost-effective technology capable of screening several genes simultaneously [4]. It is commonly used nowadays for sequencing mutated tumor genes with tumor tissue or plasma to identify and classify molecular subtypes, to address the unmet need for new drug targets in its category [5]. The mechanism of resistance to osimertinib or other third-generation EGFR-TKI was extremely complicated, and the reported results of mutation sites and/or mutation rates were much different among studies. Phenotype transformation, EGFR new point mutation, pathways activation, or targets loss were the strongest possibilities. Most studies reported that C797S mutation happened in 20–30% of patients after osimertinib initiation [6, 7]. The EGFR C797S mutation conferred resistance to third-generation EGFR-TKI. C797S mutation had been identified in cis or in trans with T790M mutation in tumor specimens from patients who experienced treatment failure with third-generation EGFR-TKIs. C797S and T790M mutation in trans were sensitive to first-generation plus third-generation EGFR-TKI but in cis they would be resistant to all [8]. Therefore, to elicit the mutated driver genes after resistance to third-generation EGFR-TKI is critically important.
2. Material and Methods
Patients enrolled in the study all had histologically confirmed metastatic lung adenocarcinoma. EGFR T790M mutation was confirmed by tumor tissue or serum, which was tested by the ARMS PCR or Next-Generation Sequencing (NGS) before osimertinib treatment. Patients were all treated with osimertinib with a dose of 80 mg oral daily after resistance to gefitinib or erlotinib treatment. Osimertinib acquired resistance was confirmed by CT or PET-CT scan according to RECIST 1.1 [9]. Core needle biopsy (CNB) guided by CT scan was performed. DNA was extracted from 15 × 5 um sliced sections of FFPE tumor tissue. Tumor area was evaluated and confirmed by pathologist. In order to ensure adequacy of sequences and mutation detection, at least 20% tumor area on each slice was set as a minimum. 10 ml blood was drawn and centrifuged for sequencing control and for germline genes mutation test. NGS was performed with HiSeq3000/HiSeq4000 Illumina techniques. 4278 exons of 288 common genes; intron, promoter, and fusion of 38 genes; and coding area of 728 genes were tested for somatic mutations. 11 germline mutations were also tested. The ultra-deep coverage of genes of interest was 1,000x for tumor tissue and 10,000x for serum.
This study was approved by the Ethics Committee of Affiliated Qingdao Central Hospital of Qingdao University, and Informed consent to reveal patients' medical history for publishing was obtained before submitting this manuscript.
3. Results
A total of 9 Chinese patients were studied. There were 5 female patients and 4 male patients, median age 66-year, range 51–89. All patients were core needle biopsied and adenocarcinomas were confirmed by pathologists; there was no SCLC transformation found. Tissues obtained sources for testing and genetic testing methodologies were listed (Table 1). There were 62 point mutations, 2 altered gene copies, 2 amplifications, and 1 EML4-ALK gene fusion found in these 9 patients (Table 2). No MET or HER2 amplification was found in this cohort study. All the 9 patients still maintained initial EGFR 19 del or L858R activating mutations; meanwhile, 7 of them kept EGFR T790M mutations. Furthermore, among the 7 patients, 5 had secondary EGFR C797S and/or C797G mutations, which all happened in the same allele with T790M mutation. Two patients had EGFR C797G 2389T>G mutations; 1 had EGFR C797S 2390G>C mutation; 1 had EGFR C797S 2389 T>A and 2390 G>C 2 points mutations; and 1 had EGFR C797S 2389 T>A, 2390 G>C 2 points and C797G 2389 T>G mutations. Subjects #1 and #5 had acquired EGFR T790M mutations tested by NGS, but lost them after osimertinib treatment. There were 9 point mutations before osimertinib treatment, which increased to 22 point mutations after resistant to osimertinib. Most pre-osimertinib point mutations could be tested after osimertinib treatment with the exception of T790M mutation and WSCD2 mutation (Table 3). The above 2 patients were treated back with gefitinib, one having had stable disease for 3 months, the other one just started. Subject #3 had a low mutation rate of EML4-ALK fusion and simultaneously had EGFR L858R, T790M mutation, and C797G mutation and was treated with crizotinib but had progression in 1 month. He was treated with pemetrexed for 2 cycles and had a confirmed partial response. Subject #6 had harboring high MAF of EGFR EXON21 (c.2500G>T, p.V834L), 19 del, TP53, T790M, and C797G mutations (91.1%, 85.5%, 85.2%, 61.5%, 36.6%, resp.) and was resistant to gefitinib combined with osimertinib but had stable disease with cisplatin plus pemetrexed chemotherapy. Subject #7 had EGFR C797S (2389 T>A, 2390 G>C) mutations and was treated with erlotinib, osimertinib, and cetuximab but had progression in 2 months and died 6 months later. Subject #8 had EGFR 19 del and T790M and C797S/G mutations and was treated with BSC and died after 6 months. The other 3 patients were in chemotherapy or BSC at last follow-up on December 15, 2017 (Figure 1).
Table 1.
Patients' characteristics.
| # | Sex | Age (yrs) | EGFR type | Tissues and genetic testing | |||
|---|---|---|---|---|---|---|---|
| Before osimertinib | After osimertinib | ||||||
| 1 | F | 62 | L858R | Lung | NGS | Pleura | NGS |
| 2 | M | 85 | 19 del | SL | ARMs-PCR | Lung | NGS |
| 3 | M | 66 | 19 del | Bone | ARMs-PCR | Lung | NGS |
| 4 | F | 79 | L858R | Lung | ARMs-PCR | Lung | NGS |
| 5 | F | 89 | 19 del | Lung | NGS | Lung | NGS |
| 6 | F | 66 | 19 del | Serum | ARMs-PCR | Bone | NGS |
| 7 | M | 56 | 19 del | Lung | ARMs-PCR | Lung | NGS |
| 8 | F | 75 | 19 del | Pleura | ARMs-PCR | Pleura | NGS |
| 9 | M | 51 | 19 del | Lung | ARMs-PCR | Lung | NGS |
| Median | 66 | ||||||
SL denotes supraclavicular lymph node.
Table 2.
Mutation types and mutant allele fraction (MAF) (%).
| #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | Rate | Mutation | Rates | Mutation | Rates | Mutation | Rates | Mutation | Rates | Mutation | Rates | Mutation | Rates | Mutation | Rates | Mutation | Rates |
| L858R | 63.2 | 19 del | 48.6 | 19 del | 2.8 | L858R | 22.2 | 19 del | 2.1 | 19 del | 85.5 | 19 del | 51.9 | 19 del | 24.3 | 19 del | 65.68 |
| IL7R | 31.5 | T790M | 15.1 | T790M | 4.6 | T790M | 2.2 | PIK3CA | 1.8 | T790M | 61.5 | T790M | 32.5 | T790M | 18.7 | T790M | 46.04 |
| TENM3 | 27.1 | JAK1 | 13.7 | C797G | 2.6 | NOTCH3 | 27.5 | TP53 | 1.7 | C797G | 36.6 | C797S T>A | 18.4 | C797S T>A | 6 | C797S G>C | 11.4 |
| CCND3 | 14.3 | NF2 | 11.4 | EML4-ALK | 0.9 | IGF1R | 15.1 | RNASEL | 1.2 | EGFR 834>L | 91.1 | C797S G>C | 0.8 | C797S G>C | 5 | TP53 | 35.62 |
| ROS1 (6737 G>A) | 2.9 | TET2 | 10.5 | PIK3CA | 0.9 | ZNF512B | 10.5 | TET2 | 0.8 | TP53 | 85.2 | EGFR2134T>A | 0.3 | C797G | 2.2 | C11orf30 | 17.1 |
| FGFR3 | 1.9 | FCGR2A | 18.1 | TMPRSS2 | 10.5 | MAP3K1 | 0.8 | TP53 | 36.7 | CTNNB1 | 38.8 | FLT4 | 12.01 | ||||
| XPC | 1.5 | MLL3 | 7.8 | RB1 | 20.5 | MED12 | 34.8 | ||||||||||
| ATM | 1.5 | PIK3CB | 6.7 | PIK3CA | 0.7 | APC | 21.9 | ||||||||||
| FAM135B | 1.2 | EGFR L406Q | 6.6 | BRCA1 | 0.6 | SCF1R | 4.6 | ||||||||||
| PALB2 | 1.1 | FLT4 | 4.9 | ALK | 0.6 | FCGR2R | 2.5 | ||||||||||
| MLL | 1 | ERBB3 | 3.3 | JAK1 | 0.5 | ||||||||||||
| ROS1 (1611A>G) | 1 | FAT1 | 3.2 | ROS1 2495T>A | 0.4 | ||||||||||||
| CDKN2A | 0.2 | HPS3 | 3.2 | ||||||||||||||
| Altered copy | GNAS | 2.5 | |||||||||||||||
| CDKN2B DEL | 0.3 | CLB | 2.2 | ||||||||||||||
| CDKN2A DEL | 0.2 | CDKN1A | 1.9 | ||||||||||||||
| SMO | 1.9 | ||||||||||||||||
| ROBO3 | 1.7 | ||||||||||||||||
| PTCH1 | 1.7 | ||||||||||||||||
| CDK12 | 1.4 | ||||||||||||||||
| MDM2 | 1.2 | ||||||||||||||||
| FOXL2 | 1.1 | ||||||||||||||||
| Amplification | |||||||||||||||||
| MDM2 | 3.8 | ||||||||||||||||
| CDK4 | 5.0 | ||||||||||||||||
Table 3.
Mutation types and mutant allele fraction (MAF) (%) changes.
| #1 | #5 | ||||||
|---|---|---|---|---|---|---|---|
| Before osimertinib | After osimertinib | Before osimertinib | After osimertinib | ||||
| Mutation | Rate | Mutation | Rates | Mutation | Rates | Mutation | Rates |
| L858R | 42.9 | L858R | 63.2 | 19 del | 9.6 | 19 del | 2.1 |
| IL7R | 19.1 | IL7R | 31.5 | T790M | 1.1 | PIK3CA | 1.8 |
| T790M | 5.7 | TENM3 | 27.1 | TP53 | 7.11 | TP53 | 1.7 |
| CCND3 | 5.8 | CCND3 | 14.3 | RNASEL | 1.2 | ||
| WSCD2 | 5.1 | ROS1 (6737 G>A) | 2.9 | TET2 | 0.8 | ||
| FGR3A | 4.1 | FGFR3 | 1.9 | MAP3K1 | 0.8 | ||
| XPC | 1.5 | ||||||
| ATM | 1.5 | ||||||
| FAM135B | 1.2 | ||||||
| PALB2 | 1.1 | ||||||
| MLL | 1 | ||||||
| ROS1 (1611A>G) | 1 | ||||||
| CDKN2A | 0.2 | ||||||
| Altered copy | |||||||
| CDKN2B DEL | 0.3 | ||||||
| CDKN2A DEL | 0.2 | ||||||
Figure 1.
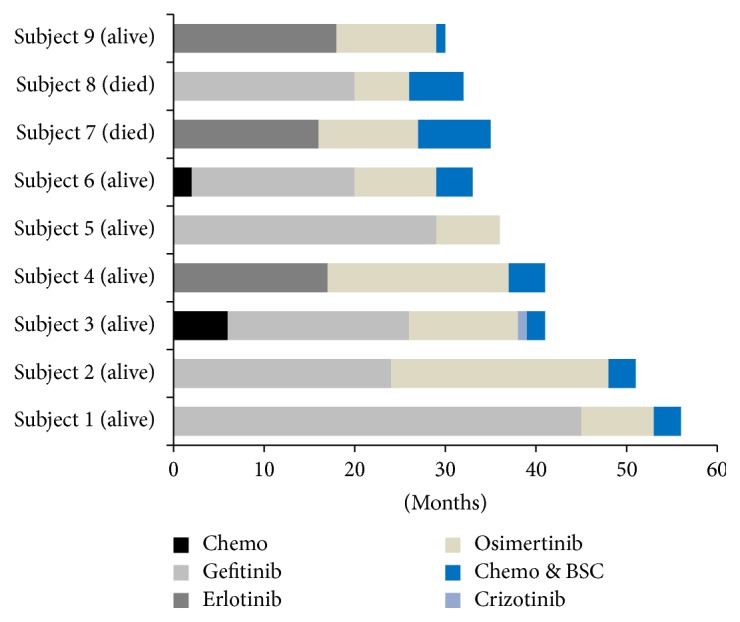
Treatment and outcomes.
4. Discussion
Activation of epidermal growth factor receptor (EGFR) triggers antiapoptotic signaling, proliferation, angiogenesis, invasion, and metastasis, which lead to development and progression of NSCLC [10]. Inhibition of EGFR by tyrosine kinase inhibitors such as gefitinib and erlotinib has increased tumor response and prolonged patients' survival. However, acquired resistance will finally happen, with a progression-free survival (PFS) of around 9–13 months [11, 12]. The substitution of threonine with methionine at amino acid position 790 (T790M), as the second mutation in EGFR, is the most common resistance mechanism and is detected in tumor cells from more than 50–60% of patients after disease progression. This mutation enhances ATP affinity and reduces the ability of ATP-competitive reversible EGFR-TKI binding to EGFR tyrosine kinase domain, which results in cancer cells resistant to gefitinib and erlotinib [1]. Osimertinib is an oral, potent, irreversible EGFR-TKI and inhibits kinase activity of EGFR sensitive mutation and T790M resistant mutation [13, 14]. Osimertinib for second-line treatment in EGFR T790M positive NSCLC was associated with a response rate of 61% and PFS of 9.6 months [13]. But the mechanism of osimertinib resistance is much complicated [6, 7]. Third-generation EGFR-TKIs (WZ 4002, rociletinib, osimertinib) have pyrimidine structure and covalent bond with EGFR Cys797 at ATP binding pocket. Cys797 point mutation (C797S/G/R) intercepts 3rd-generation EGFR-TKIs covalent binding to Cys797 at ATP binding pocket, which results in drug resistance [15, 16]. Thress et al. [17] reported results of 15 patients NGS after osimertinib progression: 6/15 cases acquired the C797S mutations, 5/15 cases maintained the T790M mutations but did not acquire the C797S mutation, and 4/15 cases lost the T790M mutations. In our results, all 5 acquired C797S/G mutations, patients still maintained EGFR T790M mutations, and C797S/G and T790M mutations were in the same allele. In in vitro study, tumor cells harboring C797S mutation without T790M mutation are still sensitive to quinazoline-based EGFR inhibitors like gefitinib or erlotinib, and, harboring EGFR C797S in trans with T790M mutations are sensitive to a combination of first- and third-generation EGFR-TKI and in cis are resistant to all current EGFR inhibitors [18, 19].
HER2 or MET amplifications were also reported in osimertinib-resistant NSCLC patients. EGFR T790M mutation and HER2 amplification appeared to be mutually exclusive while MET amplification occurred with or without EGFR T790M mutation [20, 21]. It was hypothesized that tumor heterogeneity, HER2, or MET subclones initiated an independent pathway for tumor progression [17, 22].
There were 2 patients who had acquired low MAF PIK3CA mutations; 1 patient had acquired NOTCH3, IGF1R, and PIK3B mutations. These point mutations are all associated with osimertinib use and might correlate with drug resistance [23], which needs further study to elicit new methods to overcome it.
Small-cell lung cancer transformation is rare in NSCLC patients resistant to EGFR-TKIs treatment. It might preexist before first-/second-/third-generation EGFR-TKIs treatment [24]. So, rebiopsy is critically important in the diagnosis of SCLC transformation after resistance to 3rd-generation EGFR-TKIs.
In our study, 2 patients had been tested by NGS before and after osimertinib treatment. There were 5 point mutations excluding EGFR activating mutation and EGFR T790M mutation before osimertinib treatment. After resistance to osimertinib, 16 new point mutations were found except loss of T790M mutation and WSCD2 mutation. This phenomenon indicated that the more the lines of EGFR-TKIs therapies were initiated, the more the point mutations would happen. These 2 patients were treated back with gefitinib; 1 had stable disease for 3 months. It was indicated that 1st-generation EGFR-TKI could be used in patients who lost T790M mutation but maintained EGFR 19 del or L858R activating mutation. 1 patient had low rate EML4-ALK gene fusion (0.9%), EGFR 19 del (2.8%), and T790M (4.6%) and C797G (2.6%) mutations and was treated with crizotinib and progressed in 1 month but responded to chemotherapy thereafter. It might be that tumor cells with EML4-ALK gene fusion were too low. Subjects #6 and #7 had high MAF of EGFR 19 del, T790M, and C797S or C797G mutations and were resistant to 1st- and 3rd-generation combination. So, chemotherapy is the optimal treatment for C797S/G in cis with T790M mutation before new drugs can overcome it.
In this study, 5/9 (55.6%) patients had acquired C797S/G mutations, all maintained with T790M mutation in the same allele. In these 5 patients, 3 times of c.2389T>G, 2 times of c.2389T>A, and 3 times of c.2390G>C point mutations were found. The C797S/G mutations were maintained with T790M mutation in cis, and C797S/G mutations happened to be higher than previous report [17]. In the 9 patients, there were a total of 62 point mutations, 2 altered copy numbers, and 2 amplifications, and 1 patient with 2 points C797S (2389 T>A, 2390 G>C) mutations and 1 patient had 2 points C797S and C797G mutations, which made the mechanism of resistance to osimertinib more complicated.
5. Conclusions
Genetic mutation is much complicated after osimertinib treatment in EGFR positive non-small-cell lung cancer. EGFR C797S/G mutation and the same one presented on the same allele with EGFR T790M mutation were the most common mutation feature and played a key role in resistance to osimertinib in Chinese patients with NSCLC. Tumor cells losing T790M mutation and maintaining EGFR activating mutation might be benefit from first-generation EGFR-TKI treatment. To elicit a chemical to overcome EGFR C797S/G point mutation with T790M mutation in the same allele is critically important.
Acknowledgments
The authors sincerely appreciate the superb help by Qing Yan and Ye Wang for genetic consultation at Department of Central Laboratory, the Affiliated Central Hospital of Qingdao University. They thank Professor Wendy Wang of Feinstein Institute for Medical Research and Professor John Califra in New York for helping in editing the manuscript. This study was supported by Grants from National Health and Family Planning Commission of Peoples Republic of China, Medical Technology Research Center for Health Development Grants W2012FZ007 (Youxin Ji).
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this article.
Authors' Contributions
Keke Nie and Haiping Jiang made equal contributions to this study.
References
- 1.Suda K., Onozato R., Yatabe Y., Mitsudomi T. EGFR T790M mutation: A double role in lung cancer cell survival? Journal of Thoracic Oncology. 2009;4(1):1–4. doi: 10.1097/JTO.0b013e3181913c9f. [DOI] [PubMed] [Google Scholar]
- 2.Yin Y., Li J. Osimertinib in EGFR T790M - Positive lung cancer. The New England Journal of Medicine. 2017;376(20):p. 1993. doi: 10.1056/NEJMc1703339. [DOI] [PubMed] [Google Scholar]
- 3.Knebel F. H., Bettoni F., Shimada A. K., et al. Sequential liquid biopsies reveal dynamic alterations of EGFR driver mutations and indicate EGFR amplification as a new mechanism of resistance to osimertinib in NSCLC. Lung Cancer. 2017;108:238–241. doi: 10.1016/j.lungcan.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Vanni I., Coco S., Truini A., et al. Next-generation sequencing workflow for NSCLC critical samples using a targeted sequencing approach by ion torrent PGM platform. International Journal of Molecular Sciences. 2015;16(12):28765–28782. doi: 10.3390/ijms161226129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veldore V. H., Patil S., Satheesh C., et al. Genomic profiling in a homogeneous molecular subtype of non-small cell lung cancer: an effort to explore new drug targets. Indian Journal of Cancer. 2015;52(2):243–248. doi: 10.4103/0019-509x.175843. [DOI] [PubMed] [Google Scholar]
- 6.Oztan A., Fischer S., Schrock A. B., et al. Emergence of EGFR G724S mutation in EGFR-mutant lung adenocarcinoma post progression on osimertinib. Lung Cancer. 2017;111:84–87. doi: 10.1016/j.lungcan.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Tang Z., Su M., Guo X., et al. Increased expression of IRE1α associates with the resistant mechanism of osimertinib (AZD9291)-resistant non-small cell lung cancer HCC827/OSIR cells. Anti-Cancer Agents in Medicinal Chemistry. 2017;17 doi: 10.2174/1871520617666170719155517. [DOI] [PubMed] [Google Scholar]
- 8.Hidaka N., Iwama E., Kubo N., et al. Most T790M mutations are present on the same EGFR allele as activating mutations in patients with non–small cell lung cancer. Lung Cancer. 2017;108:75–82. doi: 10.1016/j.lungcan.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz L. H., Litière S., De Vries E., et al. RECIST 1.1 - Update and clarification: From the RECIST committee. European Journal of Cancer. 2016;62:132–137. doi: 10.1016/j.ejca.2016.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J.-Q., Zhong W.-Z., Zhang G.-C., et al. Better survival with EGFR exon 19 than exon 21 mutations in gefitinib-treated non-small cell lung cancer patients is due to differential inhibition of downstream signals. Cancer Letters. 2008;265(2):307–317. doi: 10.1016/j.canlet.2008.02.064. [DOI] [PubMed] [Google Scholar]
- 11.Zhou C., Wu Y.-L., Chen G., et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. The Lancet Oncology. 2011;12(8):735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 12.Mok T. S., Wu Y.-L., Thongprasert S., et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. The New England Journal of Medicine. 2009;361(10):947–957. doi: 10.1056/nejmoa0810699. [DOI] [PubMed] [Google Scholar]
- 13.Jänne P. A., Chih-Hsin Yang J., Kim D.-W., et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. The New England Journal of Medicine. 2015;372(18):1689–1699. doi: 10.1056/nejmoa1411817. [DOI] [PubMed] [Google Scholar]
- 14.Jiang T., Zhou C. Clinical activity of the mutant-selective EGFR inhibitor AZD9291 in patients with EGFR inhibitor-resistant non-small cell lung cancer. Translational Lung Cancer Research. 2014;3(6):370–372. doi: 10.3978/j.issn.2218-6751.2014.08.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia Y., Yun C.-H., Park E., et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature. 2016;534(7605):129–132. doi: 10.1038/nature17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menon R., Müller J., Schneider P., et al. A Novel EGFRC797 Variant Detected in a Pleural Biopsy Specimen from an Osimertinib-Treated Patient Using a Comprehensive Hybrid Capture-Based Next-Generation Sequencing Assay. Journal of Thoracic Oncology. 2016;11(9):e105–e107. doi: 10.1016/j.jtho.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Thress K. S., Paweletz C. P., Felip E., et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nature Medicine. 2015;21(6):560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z., Yang J.-J., Huang J., et al. Lung Adenocarcinoma Harboring EGFR T790M and In Trans C797S Responds to Combination Therapy of First- and Third-Generation EGFR TKIs and Shifts Allelic Configuration at Resistance. Journal of Thoracic Oncology. 2017 doi: 10.1016/j.jtho.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 19.Ercan D., Choi H. G., Yun C.-H., et al. EGFR mutations and resistance to irreversible pyrimidine-based EGFR inhibitors. Clinical Cancer Research. 2015;21(17):3913–3923. doi: 10.1158/1078-0432.CCR-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelman J. A., Zejnullahu K., Mitsudomi T., et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 21.Piotrowska Z., Niederst M. J., Karlovich C. A., et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discovery. 2015;5(7):713–722. doi: 10.1158/2159-8290.CD-15-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Planchard D., Loriot Y., André F., et al. EGFR-independent mechanisms of acquired resistance to AZD9291 in EGFR T790M-positive NSCLC patients. Annals of Oncology. 2015;26(10):2073–2078. doi: 10.1093/annonc/mdv319.mdv319 [DOI] [PubMed] [Google Scholar]
- 23.Ramalingam S. S., Yang J. C., Lee C. K., et al. Osimertinib As First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. Journal of Clinical Oncology. 2017 doi: 10.1200/JCO.2017.74.7576.JCO2017747576 [DOI] [PubMed] [Google Scholar]
- 24.Minari R., Bordi P., Del Re M., et al. Primary resistance to osimertinib due to SCLC transformation: Issue of T790M determination on liquid re-biopsy. Lung Cancer. 2018;115:21–27. doi: 10.1016/j.lungcan.2017.11.011. [DOI] [PubMed] [Google Scholar]


