Figure 6. Mutations at the interaction interfaces can affect regulation of cancer‐associated processes.
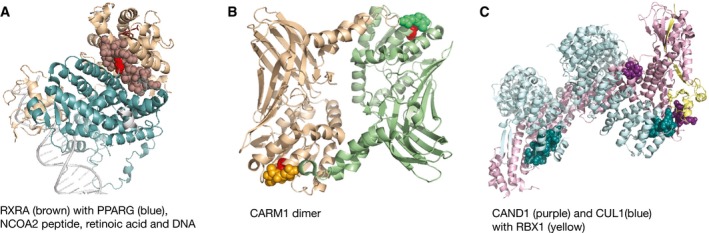
- Residues within the interaction interface of the RXRA protein (brown) toward the PPARG receptor (peroxisome proliferator‐activated receptor gamma, blue) are shown as spheres. A signal for this interface was driven by the hotspot mutation in RXRA (shown as a red sphere). Other protein regions in the representative PDB structure (3dzy) are shown as ribbons. The NCOA2 coactivator peptide is not visible from the shown angle, while the retinoic acid can be partly seen above the interface and it is represented with dark red sticks.
- CARM1 protein dimer is shown within the representative PDB structure (5dx0). Again, the interface segments are depicted as spheres and the rest of the protein as ribbons. Significant signal within the interface is driven by the CARM1 hotspot mutation that is shown in red.
- Interaction interfaces that gave a strong signal for the clustering of cancer mutations in the CAND1 (cullin associated and neddylation dissociated 1, purple) and CUL1 (culling homolog 1, blue) proteins are shown as spheres and other protein segments as ribbons. The shown PDB structure (1u6g) represents a complex that additionally contains the RBX1 protein (RING‐box protein 1, yellow).
