Figure 3.
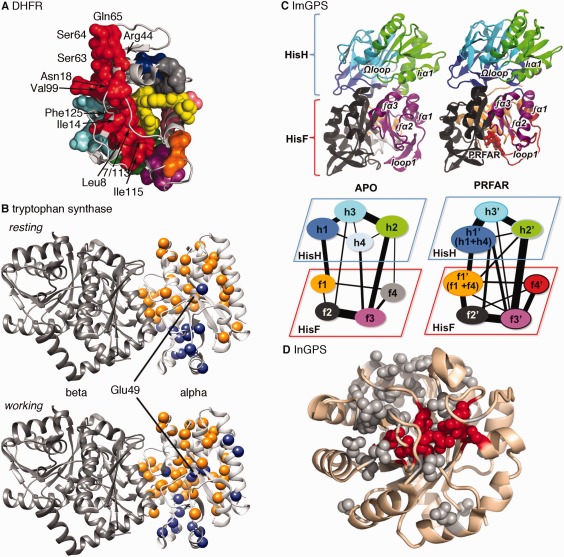
Dynamic amino acid interaction networks in the function and allosteric regulation of enzymes. A. The CONTACT approach, based on fitting alternative side‐chain conformations in X‐ray diffraction maps, was used to identify amino acid interaction networks in E. coli dihydrofolate reductase. This approach provides experimental insight into the connections between distant parts of the enzyme, including those regions identified as being part of a network of coupled promoting motions [see Fig. 1(A)]. Reprinted by permission from Macmillan Publishers Ltd.98 B. The CHESCA approach,111 based on analyzing NMR chemical shift changes induced by protein perturbations, was used to identify amino acid interactions networks in the alpha subunit of tryptophan synthase. The orange and blue spheres indicate two different networks. The catalytic residue Glu49 has a different network association in the apo resting state and in the catalytically active working state. Network residues also include those at the alpha/beta interface. C. Amino acid interaction network communities (represented by different colors) in imidazole glycerol phosphate synthase from the histidine biosynthetic pathway, which includes both HisH and HisF subunits. Ammonia is passed from the HisH to the HisF subunits. Binding of PRFAR (N’‐[(5’‐phosphoribulosyl)formimino]‐5‐aminoimidazole‐4‐carboxamide ribonucleotide) changes community interactions. Reprinted with permission from Ref. 116. D. Statistical coupling analysis, based on amino acid covariation in a multiple sequence alignment,166 was used to identify amino acid interaction networks (or sectors) in indole‐3‐glycerol phosphate synthase. Residues highlighted in red are those at the active site. Amino acid substitutions at many of these positions lead to changes in competitive effectiveness of yeast strains carrying these variants, according to high throughput mutational analyses. Reprinted with permission from Ref. 132.
