Figure 7.
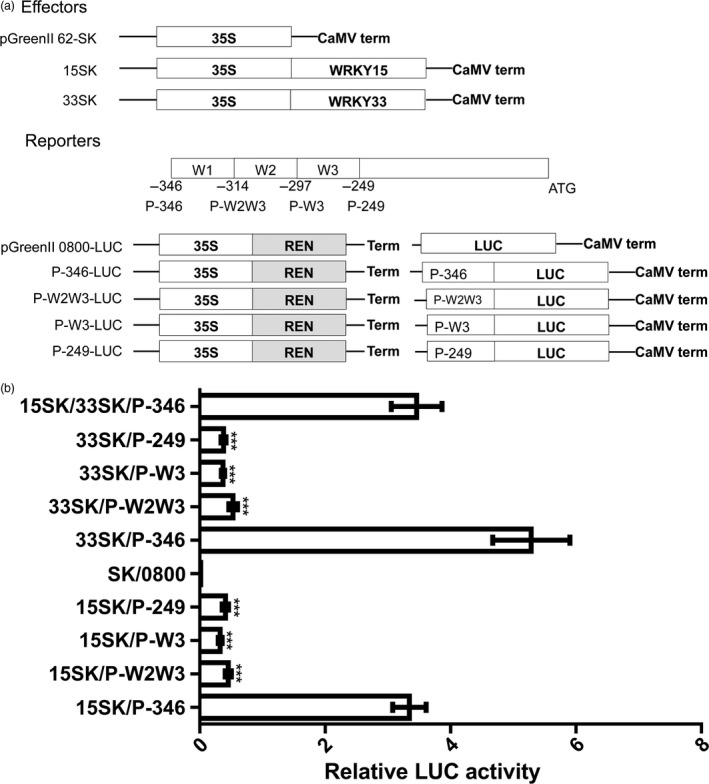
Defined region bound for activation by BnWRKY15 and BnWRKY33 obtained by Arabidopsis protoplast transient assays. (a) Schematic representation of effector and promoter reporter constructs used in Arabidopsis protoplast transient assays. The open reading frame of BnWRKY15 or BnWRKY33 was inserted into pGreenII 62‐SK to generate 15SK or 33SK plasmids as effectors. The promoter regions containing the first W‐box, second W‐box and third W‐box were designated regions W1, W2 and W3, respectively. Different promoter regions (P‐346, P‐W1W2, P‐W3 and P‐249) of BnWRKY33 fused with the firefly luciferase (LUC) gene were used as reporters. The location of each truncated promoter is indicated above the name of each promoter. Empty pGreenII 62‐SK plasmids and empty pGreenII 0800‐LUC construct were cotransformed into protoplasts, serving as a negative control. REN refers to the Renilla luciferase gene, which served as an internal control. LUC refers to the firefly luciferase gene. (b) The abilities of BnWRKY15 and BnWRKY33 to bind different promoter regions of BnWRKY33 are indicated by the relative LUC activities, which were calculated by comparing LUC activities to REN activities. The effectors and a reporter were cotransfected. P‐346, P‐W2W3, P‐W3 and P‐249 represent different reporter plasmids that contain different inserted promoter regions, as indicated in (a). SK and 0800 refer to empty effector and empty reporter plasmids. The mark on the y‐axis indicates different combinations of effector and reporter plasmids, and x‐axis represents LUC enzyme activities of these combinations in Arabidopsis protoplasts. The data represent the means ± standard errors (n ≥ 3). Statistical analyses were performed using Student's t‐test: ***P < 0.001.
