Figure 2.
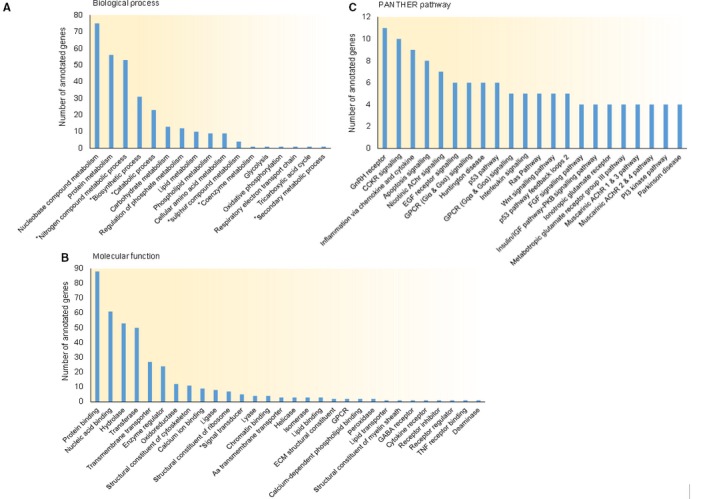
miR‐122 target profile reveals IR‐relevant ontologies and pathways. miR‐122 predicted targets were analysed for molecular function, biological process and associated pathways with PANTHER Gene Ontology Tool. (A) Biological process ontology includes protein metabolism (56), carbohydrate metabolism (13) and lipid metabolism (10). (B) Molecular function ontology revealed several IR‐relevant biochemical processes including transmembrane transport (27), calcium ion binding (9), GPCR activity (2), peroxidase activity (2) and lipid transporter activity (1). (C) PANTHER pathway analysis identified key signalling pathways including apoptosis signalling (8), EGFR signalling (6), GPCR signalling (11), Ras pathway (5), insulin/IGF pathway‐PKB signalling (4) and PI3 kinase pathway (4). *Indicates a general level 1 ontology in (B) and level 2 ontology in (A), all other listed ontologies are more specific terms at either level 2 in (B) or level 3 in (A). DIANA‐microT‐CDS predictions for miR‐122 are provided in Data S1. PANTHER results with ontologies and corresponding gene lists are provided in Data S2
