Abstract
Despite the development of temozolomide (TMZ), a novel type of glioma therapeutic drug, malignant glioma remains to cause severe damage to human health. The present study aimed to utilize the molecular biological differences between tumor and normal cells to design TMZ derivatives with improved selectivity and targeting using computer-aided drug design (CADD). Taking alkylglycerone phosphate synthase (AGPS) as a target, a 3D structure-activity relationship model was built using CADD technology; molecular docking of isothiocyanate (ITC) and TMZ compounds was conducted; ITC-TMZ derivatives were designed; and predictions on the absorption, distribution, metabolism and excretion (ADME) processes and toxicity of the ITC-TMZ derivatives were established in order to obtain improved understanding of the structure-activity relationship of the candidate compounds. Using these techniques, it was identified that the docking scores of the structural derivatives S1-9 were higher than that of TMZ. Additionally, S3, −6, −7, −8, −9 and −10 exhibited enhanced ADME and similar toxicity to that of TMZ. The half maximal inhibitory concentrations of the CADD derivatives were also assessed in the glioma U87MG and U251 cell lines, and the activities of S1, −3, −8 and −10 were determined to be greater than that of TMZ, suggesting their potential as anti-cancer drugs with adequate AGPS targeting, ADME/toxicity and anti-tumor activity.
Keywords: glioma, temozolomide, computer-aided drug design, alkylglycerone phosphate synthase
Introduction
Malignant glioma may cause severe damage to human health, and accounts for ~70% of primary malignant brain tumors, with an annual incidence of ~5/100,000 in China (1). Also in China, the number of new cases exceeds 14,000 each year (2). For patients with grade IV glioblastoma according to World Health Organization criteria (3), median survival time is between 14 and 17 months and total annual mortalities may reach 30,000 worldwide (4). Temozolomide (TMZ) is a novel type of glioma therapeutic drug, which is able to penetrate the blood-brain barrier to exert targeted therapeutic effects on brain tumor tissue (5,6). To date, studies have aimed to enhance the efficacy and reduce the toxic side effects of TMZ by reconstructing its chemical structure (7,8). To build on these previous studies, the present study utilized the molecular biological differences between tumor and normal cells to design TMZ derivatives with enhanced selectivity and targeting activities through rational computer-aided drug design (CADD).
Alkylglycerone phosphate synthase (AGPS) is considered to serve as an oncogene through its involvement in lipid metabolic processes associated with cancer pathogenicity (9). A previous study by our group demonstrated through RNA interference technology that silencing of AGPS expression in glioma and liver cancer cells inhibited the proliferation and invasion of the tumor cells, and improved the drug sensitivity of resistant cells (10). Thus, AGPS may be a therapeutic candidate for glioma therapy.
CADD may overcome the disadvantages of traditional drug development processes, including their long clinical trial cycle period, low combinatorial variety and low efficiency, by simulating the interaction between drugs and receptors of biological macromolecules (11,12). CADD has identified numerous drugs to date, and is becoming a core technology in the field of novel drug development (13). In particular, it may aid to identify more optimized TMZ derivatives with enhanced therapeutic effect in glioma patients.
On the basis of previous study (14), the current study has taken AGPS as a target, built a 3D structure-activity relationship model through CADD technology, and conducted molecular docking of isothiocyanate (ITC)-TMZ compounds. A total of 10 ITC-TMZ derivatives were identified as novel compounds with adequate targeting of AGPS. Additionally, predictions on the absorption, distribution, metabolism and excretion (ADME) processes and toxicity of the ITC-TMZ derivatives were established, along with anti-tumor activity in vitro, in order to elucidate the structure-activity relationship of the candidate compounds, and thus further optimize the structure of the lead compounds.
Materials and methods
Protein structure database
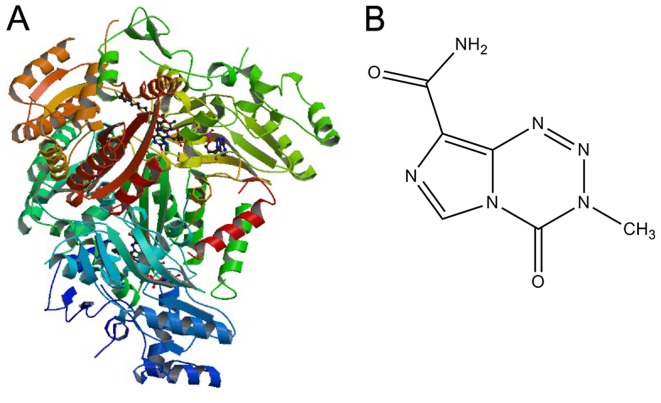
A 3D structure model of AGPS (Fig.1A) was downloaded from the Protein Data Bank (www.rcsb.org; PDB ID: 2UUV) and the protein modules were processed using Discovery Studio 3.5 software (BIOVIA, San Diego, CA, USA) as follows: A single chain (the A chain) in the tetramer was retained; missing atoms to residues were added; the loop area was repaired; combinations and free water molecules were removed; unsaturated atoms were hydrogenated; valence was repaired; protein and original ligands were retained; and six basic pharmacophore elements were selected, namely hydrogen bond acceptor, hydrogen bond donor, hydrophobic, positive electricity center, negative electricity center and aromatic center. This obtained a final 3D AGPS structure for docking. The structure of TMZ used is depicted in Fig. 1B (15).
Figure 1.
Structures of AGPS and TMZ. (A) 3D structural model of AGPS; (B) structure of TMZ. TMZ, temozolomide; AGPS, alkylglycerone phosphate synthase.
Virtual screening
The structures of TMZ derivatives were combined with benzyl isothiocyanate (BITC) and the ITC-TMZ derivatives were drawn using ChemBioOffice 2010 (PerkinElmer, Waltham, MA, USA), and each structure was saved in mol2 format. The structures were imported into Discovery Studio 3.5 for virtual screening. The CDOCKER protocol in Discovery Studio 3.5 was used for molecular docking; ‘CDOCKER energy’ was used as a scoring index, with higher scores indicating greater docking affinity.
The experiment screened 50 molecules
Firstly, it used the pharmacophore model generated by the HipHop algorithm to perform fit value sorting of the compounds based on the characteristics of the pharmacophore model, with small molecular compounds with the highest scores retained (16). The pharmacophore model generated by the Counting Belief Propagation algorithm was then used on reserved compounds to obtain preferable compounds that conformed to the characteristics of the two pharmacophore types (17). BITC was used as a control for the virtual screening.
ADME and toxicity prediction
Protein structure processing was completed with the QikProp module of Schrodinger Suite 2009 software (Schrodinger, LLC, Cambridge, MA, USA). The downloaded AGPS target crystal structure (PDB ID: 2UUV) was imported into the Schrodinger software and processed as follows: Metal ions were removed; unsaturated atoms were hydrogenated; amino acid residues at the N and C termini were made neutral; and protein and original ligands were retained to remove atom and water molecule impurities. The OPLS_2005 position was selected to optimize protein structure and utilize sample water orientations as parameters to optimize hydrogen bonding.
Ligand preparation utilized ChemBioDraw Ultra 11.0 in the ChemBioOffice 2010 to draw planar structures of the micromolecules, which were imported into ChemBio3D Ultra 11.0 for the generation of 3D structures, which were saved in mol2 format. The mol2fileswere then imported into Schrodinger Suite 2009 software and processed with the LigPrep module, complete desalting, add charge and produce ionization consistent with human body pH. The OPLS_2005 position was also selected for the optimized molecules to match that of the optimized acceptor. ADME parameters, namely oil-water partition coefficient (logPo/w), polarization surface area (PSA), water solubility (logS) and apparent Maden Darby Canine Kidney cell permeability (PMDCK), and toxicity factors, namely rodent carcinogenicity, mutagenicity, skin irritancy, ocular irritation and aerobic biodegradability, were subsequently simulated in the Schrodinger Suite 2009 software.
Cell lines and culture
The human glioma cell lines U87MG and U251 were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. It is important to note that U87MG has been identified to differ from the original cell line, and is probably an authentic human glioblastoma cell line of unknown origin according to DNA profiling (18).
MTS assay
A total of 5×103 cells/well were cultured in 96-well plates at 37°C with 5% CO2 for 24 h. ITC-TMZ derivatives were designed and synthesized via CADD (14) and added to the medium at final concentrations of 0 (negative control), 1, 2, 5, 10, 20, 50 and 100 µM, respectively. Subsequently, the cells were cultured for 72 h at 37°C, after which the medium was replenished with 200 µl plasma-free medium and 40 µl MTS, followed by incubation at 37°C for another 4 h. The optical density (OD) value was measured using an enzyme-labeled meter at 490 nm. Inhibition rate (%) was calculated as follows: (1-OD value of the medicine-treated group/OD value of the negative control group) × 100%. Half maximal inhibitory concentrations (IC50s) were subsequently determined using GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Virtual screening and core hopping
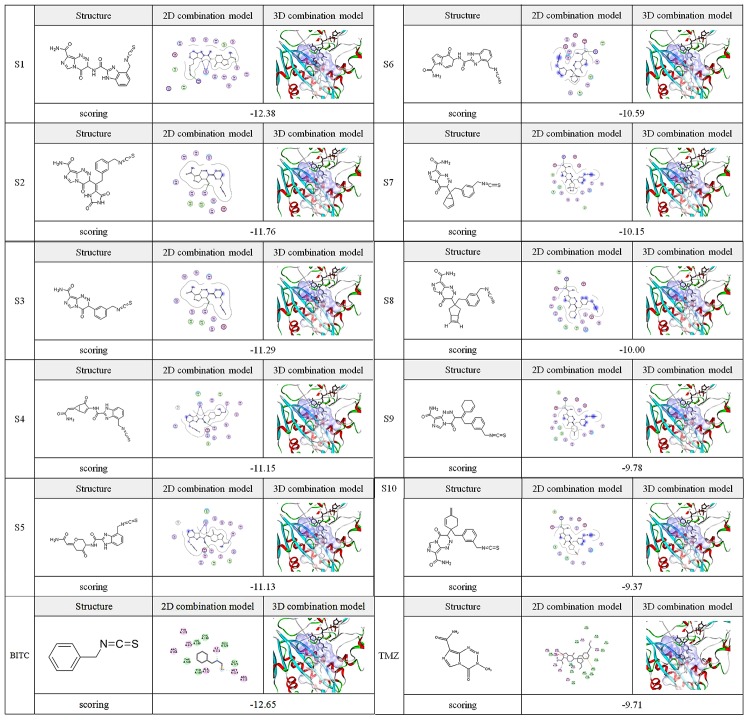
Visual screening of 50 small molecular compounds was performed using Schrodinger Suite 2009. A total of 10 ITC-TMZ derivatives and the compound BITC exhibited micromolecular structures with improved combination with the target protein AGPS compared with TMZ, determined by referring to the combination mode of the micromolecules and surrounding amino acids, and evaluation of scoring function from CDOCKER energy. The scoring of the docking between these micromolecules [structures (S)1-10 and BITC] and the AGPS active site is presented in Fig. 2. The 11 candidate compounds exhibited marked interactions with the protein cavity, with key amino acid residues forming hydrogen bonds and the hydrophobic effect occurring with nitrogen and sulfur atoms and carbon-sulfur covalent bonds of the ITC-TMZ derivative molecules. The other substituent residues in the micromolecules formed certain hydrogen bonding interactions with surrounding residues to an extent. As a result, as evident in the pattern diagrams (Fig. 2), the main ITC active group and small regions of substitute residues in the TMZ derivatives with some impurity atoms were respectively connected to both sides of the hydrophobic and rigid ring planes. These structural data suggest improved interactions and combination of the ITC-TMZ derivatives with the AGPS target compared with TMZ. Indeed, the docking scores of S1-9 and BITC were increased compared with that of TMZ, suggesting enhanced AGPS targeting.
Figure 2.
Structures, combination models with alkylglycerone phosphate synthase target and docking scores of isothiocyanate-temozolomide derivatives (S1-10). S, structural derivative; BITC, benzyl isothiocyanate.
ADME and toxicity predictions
ADME properties, including logPo/w, PSA, logS and PMDCK, and toxicity properties, including rodent carcinogenicity, mutagenicity, skin irritancy, ocular irritation and aerobic biodegradability, aid to determine whether therapeutic candidates are of clinical standard (16). The present results on ADME (Table I) and toxicity (Table II) parameters indicated the 10 ITC-TMZ derivatives to have improved interaction with AGPS. Firstly, the results suggested adequate ADME parameters for BITC regarding physico-chemical properties. In turn, the ADME parameters of S3, −6, −7, −8, −9 and −10 were optimized compared with TMZ, particularly regarding logPo/w, logS and PMDCK, suggesting the structure of BITC may improve TMZ physico-chemical properties. Meanwhile, the general toxicity parameters of the above compounds were not increased according to the predictions.
Table I.
ADME predictions of isothiocyanate-TMZ derivatives.
| ADME parameter | ||||
|---|---|---|---|---|
| Candidate | PSAa | logPo/wb | logSc | PMDCKd |
| S1 | 204.93 | 0.596 | −4.40485 | 13.345 |
| S2 | 205.35 | 1.243 | −5.29240 | 10.342 |
| S3 | 147.15 | 2.015 | −4.10003 | 111.705 |
| S4 | 162.39 | 0.869 | −4.41749 | 46.410 |
| S5 | 171.62 | 0.896 | −4.14306 | 50.200 |
| S6 | 167.32 | 1.560 | −4.32601 | 70.105 |
| S7 | 147.15 | 2.536 | −4.94407 | 183.872 |
| S8 | 147.15 | 2.99 | −5.45929 | 481.063 |
| S9 | 147.15 | 3.623 | −6.24712 | 121.998 |
| S10 | 147.15 | 3.293 | −5.66361 | 875.571 |
| BITC | 44.45 | 3.321 | −2.5125 | 10,000.000 |
| TMZ | 79.92 | −1.810 | −1.1118 | 66.744 |
Parameter reference ranges:
7.0–200.0
−2.0–6.5
−6.5–0.5
<25 indicates poor suitability and >500 indicates high suitability for drug development (16). ADME, absorption, distribution, metabolism and excretion; PSA, polarization surface area; logPo/w, oil-water partition coefficient; logS, water solubility; PMDCK, apparent Maden Darby Canine Kidney cell permeability; TMZ, temozolomide; S, structural derivative; BITC, benzyl isothiocyanate.
Table II.
Toxicity prediction of isothiocyanate-TMZ derivatives.
| Candidate | Mouse female NTP prediction | Mouse male NTP prediction | Rat female NTP prediction | Rat male NTP prediction | Ames test prediction | Skin irritancy | Ocular irritancy | Aerobic biodegradability prediction |
|---|---|---|---|---|---|---|---|---|
| S1 | NC | NC | NC | NC | NM | Mild | Moderate | ND |
| S2 | NC | NC | NC | NC | NM | Mild | Moderate | ND |
| S3 | NC | NC | NC | NC | NM | Mild | Moderate | ND |
| S4 | NC | NC | NC | NC | NM | Mild | Moderate | ND |
| S5 | NC | NC | NC | NC | NM | Mild | Moderate | ND |
| S6 | NC | NC | NC | NC | NM | Mild | Moderate | ND |
| S7 | NC | NC | NC | NC | NM | Mild | Moderate | ND |
| S8 | NC | NC | NC | NC | NM | Mild | Moderate | ND |
| S9 | NC | NC | NC | NC | NM | Mild | Moderate | ND |
| S10 | NC | NC | NC | NC | NM | Mild | Mild Severe | ND |
| TMZ | NC | NC | NC | NC | NM | None | Mild | ND |
| BITC | NC | NC | C | C | NM | Mild | Severe | ND |
NTP, National Toxicology Program; NC, non-carcinogen; C, carcinogen; NM, non-mutagen; ND, non-degradable; TMZ, temozolomide; S, structural derivative; BITC, benzyl isothiocyanate.
Anti-tumor activity of ITC-TMZ derivatives
MTS assay was performed to observe the effect of the ITC-TMZ derivatives on the viability of human glioma U87MG and U251 cells in vitro. On determination of IC50 values, S1, −3, −8 and −10 were identified to exert greater inhibitory effect on glioma cell viability compared with TMZ (Table III).
Table III.
IC50 values of isothiocyanate-TMZ derivatives in human glioma cell lines.
| IC50 (µM) | ||
|---|---|---|
| Candidate | U87MG | U251 |
| S1 | 21.7 | 28.3 |
| S2 | 35.8 | 47.4 |
| S3 | 24.5 | 31.2 |
| S4 | 115.2 | 168.5 |
| S5 | 76.7 | 62.5 |
| S6 | 98.6 | 87.1 |
| S7 | 87.4 | 88.5 |
| S8 | 41.8 | 32.2 |
| S9 | 157.6 | 91.5 |
| S10 | 45.5 | 35.8 |
| BITC | 15.2 | 18.7 |
| TMZ | 54.5 | 37.5 |
IC50, half maximal inhibitory concentration; TMZ, temozolomide; S, structural derivative; BITC, benzyl isothiocyanate.
Discussion
In 1920, cancer was described as a metabolic disease by Otto Warburg, due to his identification of the high glycolytic rate of tumor cells (19). A subsequent study demonstrated that the process of tumor development is accompanied by abnormal lipid metabolism (20). Notably, inactivation of the key enzyme for ether lipid synthesis, AGPS, has been reported to reduce ether lipid levels in tumor cells and thus decrease cancer pathogenicity (10). Additionally, its overexpression may enhance the ether lipid levels of tumor cells, increase the survival and motility of tumor cells, and promote the growth and invasion of tumors (21). Therefore, AGPS is considered to be a novel target of antitumor drugs, with specific inhibitors expected to have marked advantages over traditional chemotherapy methods. The characteristic elements of an effective inhibitor include the following: A hydrogen bond acceptor, a hydrogen bond donor, hydrophobicity, a positive electricity center, a negative electricity center and an aromatic center (22,23). The present study identified elevated docking scores for S1-9 compared with TMZ, suggesting enhanced AGPS targeting over TMZ.
ADME and toxicity are considered as important drug properties (24). In previous years, drug discovery principally focused on the discovery of active compounds. However, prior to the development stage, this method encountered problems regarding pharmacokinetics (PK), toxicity, solubility and stability (25). A previous study reported that drug failure in the developmental stage typically occurred due to poor biopharmaceutical properties (PK and bioavailability) (26). Due to high development costs, this failure results in major economic loss in the field of drug development. Therefore, it may be concluded that drug ADME and toxicity are now key considerations in the process of drug discovery, and may be used to guide the selection and optimization of lead compounds.
The present study identified that while the ADME parameters of TMZ were suitable, those of BITC were enhanced regarding physico-chemical properties, thus implicating BITC as a favorable modification for TMZ. Additionally, the physical and chemical ADME parameters of S3, −6, −7, −8, −9 and −10 were optimized compared with TMZ, particularly regarding logPo/w, logS and PMDCK. The thiocyanate group may serve a key role in these improvements. However, all of the derivatives exhibited similar toxicity to TMZ, suggesting the thiocyanate group is unable to decrease the toxicity of TMZ.
Nevertheless, the ADME parameters of S3, −6, −7, −8, −9 and −10 indicated the suitability of these candidates as potential clinical drugs, which thus require further study. Additionally, the present study investigated the activities of S1-10 in vitro following their synthesis, and identified that the ITC-TMZ derivatives S1, −8 and −10 suppressed the proliferation of U87MG and U251 cells to a greater extent than TMZ, again indicating their potential as anti-cancer drugs. Due to the established anti-tumor activity of TMZ, the thiocyanate group may also be key to proliferation inhibition.
In conclusion, the present study identified the ITC-TMZ derivatives S1, −8 and −10 to be potential anti-cancer drugs. The results for S8 and −10 were in accordance with the predictions by CADD, while S1 exhibited more optimal activity than predicted from docking score. Thus, the thiocyanate group may be involved in a complex mechanism influenced by differences in simulated and in situ environments, compound stability and cell membrane interference, among other factors, which warrants further investigation. Additionally, although U87MG was implicated to be of unknown origin, albeit an authentic human glioblastoma cell line, the results indicated the anti-tumor activity of S1, −3, −8 and −10 against glioblastoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 31501159 and 81702637), the Tianjin Public Health Key Research Project (grant no. 15KG108), the Tianjin Science and Technology Key Project on Chronic Diseases Prevention and Treatment (grant no. 16ZXMJSY00020), the Natural Science Foundation of Tianjin (grant no. 16JCQNJC12100) and the Special Program of Talent Development for Excellent Youth Scholars in Tianjin, China (grant no. TJTZJH-QNBJRC-2-9).
Availability of data and materials
All data and materials relevant to this study are described in this published article or available from the corresponding author on reasonable request.
Authors' contributions
BY was responsible for cell experiments, XL for computer-aided drug design, LH for the MTS assay and YZ for the study design.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhang H, Nie W, Zhang X, Zhang G, Li Z, Wu H, Shi Q, Chen Y, Ding Z, Zhou X, et al. NEDD4-1 regulates migration and invasion of glioma cells through CNrasGEF ubiquitination in vitro. PLoS One. 2013;8:e82789. doi: 10.1371/journal.pone.0082789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahlert UD, Nikkhah G, Maciaczyk J. Epithelial-to-mesenchymal(-like) transition as a relevant molecular event in malignant gliomas. Cancer Lett. 2013;331:131–138. doi: 10.1016/j.canlet.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 3.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Y, Yang P, Wang Q, Hu J, Xue J, Li G, Zhang G, Li X, Li W, Zhou C, et al. The effect of CXCR4 silencing on epithelial-mesenchymal transition related genes in glioma U87 cells. Anat Rec (Hoboken) 2013;296:1850–1856. doi: 10.1002/ar.22821. [DOI] [PubMed] [Google Scholar]
- 5.Gai XJ, Wei YM, Tao HM, An DZ, Sun JT, Li BS. Comparison of long-term survival between temozolomide-based chemoradiotherapy and radiotherapy alone for patients with low-grade gliomas after surgical resection. Onco Targets Ther. 2016;9:5117–5121. doi: 10.2147/OTT.S108989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashby LS, Smith KA, Stea B. Gliadel wafer implantation combined with standard radiotherapy and concurrent followed by adjuvant temozolomide for treatment of newly diagnosed high-grade glioma: A systematic literature review. World J Surg Oncol. 2016;14:225. doi: 10.1186/s12957-016-0975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Коbylinska LI, Klyuchivska OY, Grytsyna II, Finiuk N, Panchuk RR, Starykovych MO, Lehka L, Lesyk RB, Zіmenkovsky BS, Stoika RS. Differential pro-apoptotic effects of synthetic 4-thiazolidinone derivative Les-3288, doxorubicin and temozolomide in human glioma U251 cells. Croat Med J. 2017;58:150–159. doi: 10.3325/cmj.2017.58.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen TC, Cho HY, Wang W, Wetzel SJ, Singh A, Nguyen J, Hofman FM, Schönthal AH. Chemotherapeutic effect of a novel temozolomide analog on nasopharyngeal carcinoma in vitro and in vivo. J Biomed Sci. 2015;22:71. doi: 10.1186/s12929-015-0175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piano V, Benjamin DI, Valente S, Nenci S, Marrocco B, Mai A, Aliverti A, Nomura DK, Mattevi A. Discovery of Inhibitors for the Ether Lipid-Generating Enzyme AGPS as Anti-Cancer Agents. ACS Chem Biol. 2015;10:2589–2597. doi: 10.1021/acschembio.5b00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Y, Liu XJ, Yang P, Zhao M, Lv LX, Zhang GD, Wang Q, Zhang L. Alkylglyceronephosphate synthase (AGPS) alters lipid signaling pathways and supports chemotherapy resistance of glioma and hepatic carcinoma cell lines. Asian Pac J Cancer Prev. 2014;15:3219–3226. doi: 10.7314/APJCP.2014.15.7.3219. [DOI] [PubMed] [Google Scholar]
- 11.Cai Z, Zhang G, Zhang X, Liu Y, Fu X. Current insights into computer-aided immunotherapeutic design strategies. Int J Immunopathol Pharmacol. 2015;28:278–285. doi: 10.1177/0394632015588765. [DOI] [PubMed] [Google Scholar]
- 12.Scotti L, Scotti MT. Computer Aided Drug Design Studies in the Discovery of Secondary Metabolites Targeted Against Age-Related Neurodegenerative Diseases. Curr Top Med Chem. 2015;15:2239–2252. doi: 10.2174/1568026615666150610143510. [DOI] [PubMed] [Google Scholar]
- 13.Baig MH, Ahmad K, Rabbani G, Danishuddin, Choi I. Danishuddin and Choi I: Computer aided drug design and its application to the development of potential drugs for neurodegenerative disorders. Curr Neuropharmacol. 2017 Oct 16; doi: 10.2174/1570159X15666171016163510. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, Liu A, Zhang X, Qi L, Zhang L, Xue J, Liu Y, Yang P. The effect of benzyl isothiocyanate and its computer-aided design derivants targeting alkylglycerone phosphate synthase on the inhibition of human glioma U87MG cell line. Tumour Biol. 2015;36:3499–3509. doi: 10.1007/s13277-014-2986-6. [DOI] [PubMed] [Google Scholar]
- 15.Hvizdos KM, Goa KL. Temozolomide. CNS Drugs. 1999;12:237–243. doi: 10.2165/00023210-199912030-00006. [DOI] [Google Scholar]
- 16.Zhang J, Hao QQ, Liu X, Jing Z, Jia WQ, Wang SQ, Xu WR, Cheng XC, Wang RL. Molecular docking, 3D-QSAR and structural optimization on imidazo-pyridine derivatives dually targeting AT1 and PPARg. Oncotarget. 2017;8:25612–25627. doi: 10.18632/oncotarget.15778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Y, Jin YY, Wang YL, Wang RL, Lu XH, Kong DX, Xu WR. The discovery of a novel and selective inhibitor of PTP1B over TCPTP: 3D QSAR pharmacophore modeling, virtual screening, synthesis, and biological evaluation. Chem Biol Drug Des. 2014;83:697–709. doi: 10.1111/cbdd.12283. [DOI] [PubMed] [Google Scholar]
- 18.Allen M, Bjerke M, Edlund H, Nelander S, Westermark B. Origin of the U87MG glioma cell line: Good news and bad news. Sci Transl Med. 2016;8:354re3. doi: 10.1126/scitranslmed.aaf6853. [DOI] [PubMed] [Google Scholar]
- 19.Seyfried TN. Cancer as a mitochondrial metabolic disease. Front Cell Dev Biol. 2015;3:43. doi: 10.3389/fcell.2015.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liegel RP, Ronchetti A, Sidjanin DJ. Alkylglycerone phosphate synthase (AGPS) deficient mice: Models for rhizomelic chondrodysplasia punctate type 3 (RCDP3) malformation syndrome. Mol Genet Metab Rep. 2014;1:299–311. doi: 10.1016/j.ymgmr.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y, Li WM, Zhang L, Xue J, Zhao M, Yang P. Inhibitory effect of isothiocyanate derivant targeting AGPS by computer-aid drug design on proliferation of glioma and hepatic carcinoma cells. Int J Clin Exp Pathol. 2015;8:812–817. [PMC free article] [PubMed] [Google Scholar]
- 22.Turku A, Borrel A, Leino TO, Karhu L, Kukkonen JP, Xhaard H. A pharmacophore model to discover OX1 and OX2 orexin receptor ligands. J Med Chem. 2016;59:8263–8275. doi: 10.1021/acs.jmedchem.6b00333. [DOI] [PubMed] [Google Scholar]
- 23.Medeiros Turra K, Pineda Rivelli D, Berlanga de Moraes Barros S, Mesquita Pasqualoto KF. Constructing and Validating 3D-pharmacophore Models to a Set of MMP-9 Inhibitors for Designing Novel Anti-melanoma Agents. Mol Inform. 2016;35:238–252. doi: 10.1002/minf.201600004. [DOI] [PubMed] [Google Scholar]
- 24.Battu MB, Chandra AM, Sriram D, Yogeeswari P. Pharmacophore-based 3DQSAR and molecular docking studies to identify new non-peptidic inhibitors of cathepsin S. Curr Med Chem. 2014;21:1910–1921. doi: 10.2174/09298673113206660275. [DOI] [PubMed] [Google Scholar]
- 25.Chakrabarti S, Michor F. Pharmacokinetics and Drug Interactions Determine Optimum Combination Strategies in Computational Models of Cancer Evolution. Cancer Res. 2017;77:3908–3921. doi: 10.1158/0008-5472.CAN-16-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valasani KR, Chaney MO, Day VW, Shidu Yan S. Acetylcholinesterase inhibitors: Structure based design, synthesis, pharmacophore modeling, and virtual screening. J Chem Inf Model. 2013;53:2033–2046. doi: 10.1021/ci400196z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials relevant to this study are described in this published article or available from the corresponding author on reasonable request.