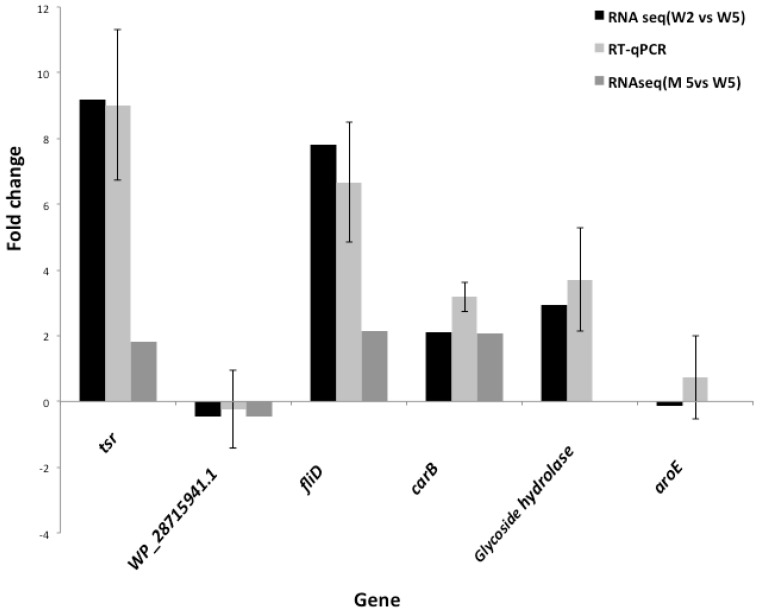
Figure 2.
RT-qPCR validation of RNA-sequencing (RNA-seq) data using six selected genes and wild-type, P. ananatis LMG 2665T RNA samples. The fold change in gene expression levels between sampling points, before Quorum Sensing (QS) (wild-type strain at OD600 = 0.2) and in the presence of QS (wild-type strain at OD600 = 0.5) were calculated using the comparative cycle threshold (CT) method [24] and data normalization was done using the ffh gene. The RT-qPCR results indicate fold change as cells shifted from before quorum sensing (OD600 = 0.2) to during quorum sensing (OD600 = 0.5) in LMG 2665T. The fold change in RNA-seq data were calculated using 2(log2FoldChange). Error bars represent the range of relative expression calculated using 2−(ΔΔCT ± StandardDeviation). Triplicates were used per biological sample. The genes aroE and the one encoding for glycoside hydrolase were not differentially expressed in RNA-seq data (M5 versus W5).