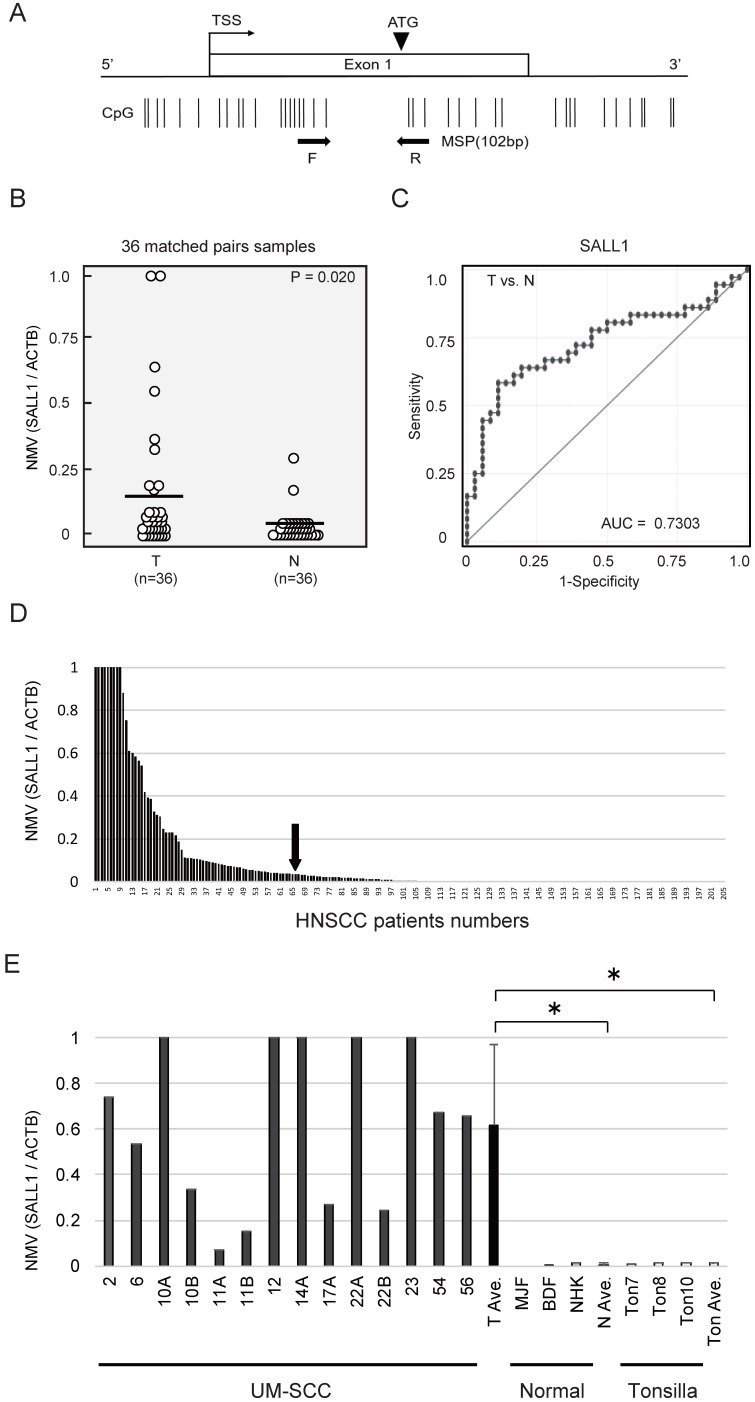
Figure 1.
Schematic representation of SALL1 gene methylation analysis by quantitative methylation-specific polymerase chain reaction (qMSP) and evaluation of SALL1 methylation in head and neck squamous-cell carcinoma (HNSCC) patients and UM-SCC cell lines. (A) SALL1 exon structure and CpG sites within expanded views of the promoter region relative to the transcript start site (TSS). Vertical lines, individual CpG sites. Straight arrows, relative location of the primers used for qMSP; bent arrow, TSS; arrowhead, translation start site (ATG). Straight arrows, relative location of the forward (F) and reverse (R) primers used for qMSP. (B) SALL1 normalized methylation values (NMVs) of head and neck tumors were higher than those of adjacent normal mucosal tissues (P = 0.020). (C) AUROC value for SALL1 was 0.7303. At the cutoff value of 0.036, sensitivity was 58.3% and specificity was 88.9%. (D) Two hundred and five DNA samples from untreated primary tumors were tested with qMSP. The cutoff NMVs (0.036) are indicated by straight arrows. (E) Representative examples of qMSP of SALL1 in eight UM-SCC cell lines, fibroblasts (MJF and BDF), keratinocytes (NHK), and normal tonsillar tissues (Ton7, Ton8, and Ton10). * P < 0.01.