Abstract
Objective: Although preoperative chemoradiotherapy (CRT) followed by total mesorectal excision (TME) is the standard treatment for locally advanced rectal cancer (LARC), the clinical efficacy differs among patients. This study was conducted to determine the association between genetic variations in the PI3K/PTEN/AKT/mTOR pathway and clinical outcomes in LARC patients.
Methods: Sixteen tagging single-nucleotide polymorphisms (SNPs) in five core genes (PIK3CA, PTEN, AKT1, AKT2, and FRAP1) were genotyped. The associations of these SNPs with tumor response to preoperative CRT, postoperative disease-free survival (DFS) and overall survival (OS) were identified. Crude odds ratios (ORs) and hazard ratios (HRs) were adjusted by age, sex, clinical stage, tumor differentiation, tumor location, cycles of preoperative chemotherapy and time interval from CRT completion to surgery.
Results: In an analysis of 97 LARC patients, the G/T+G/G genotype of AKT1:rs2498804 was associated with an increased tumor response rate (adjusted OR = 2.909, 95% confidence interval (CI), 1.127-7.505, P = 0.027). At a median of 65.7 months of follow-up, the G/C+C/C genotype of AKT2:rs8100018 was associated with a reduced risk of postoperative recurrence (adjusted HR = 0.414; 95% CI, 0.187-0.914, P = 0.029). Patients carrying the G/C+C/C genotype in AKT2:rs8100018 presented a higher 5-year DFS rate than those with the wild-type genotype (79.2% vs. 62.3%, P = 0.038). None of the SNPs were significantly associated with pathological complete response (pCR) or 5-year OS.
Conclusions: The current study indicates that genetic variations within the PI3K/ PTEN/AKT/mTOR signaling pathway are associated with the clinical outcomes of LARC patients undergoing preoperative CRT followed by radical surgery.
Keywords: rectal cancer, preoperative chemoradiotherapy, PI3K, clinical outcomes
Introduction
In recent years, rectal cancer has become a leading cause of cancer-related deaths, both in China and worldwide 1,2. As per current clinical practice guidelines, preoperative chemoradiotherapy (CRT) followed by total mesorectal excision (TME) represents the standard treatment for locally advanced rectal cancer (LARC) 3,4. Following preoperative CRT, variable responses were observed among individuals in our previous studies, as 20.9% to 42.2% of patients achieved a complete pathological response (pCR), while approximately 25% of patients presented a poor tumor response 5-7. A poor response to preoperative CRT is correlated with poor long-term oncological outcomes in these patients 8,9. Moreover, we previously observed that a proportion of patients who received preoperative CRT and surgery experienced treatment-related toxicity and surgical complications including leucopenia, thrombocytopenia, radiation proctitis, anastomotic leakage and poor wound healing 10,11. At present, the treatment responses and long-term outcomes of rectal cancer patients treated with preoperative CRT are often predicted by clinicopathological factors, such as tumor-node-metastasis (TNM) stage, tumor differentiation, location, and carcinoembryonic antigen (CEA) level 12,13. However, these clinical factors are influenced by estimates and measurement techniques and do not accurately predict treatment outcomes. Therefore, it is necessary to identify biomarkers that can precisely predict the therapeutic response and oncologic prognosis to optimize individual treatment.
Recent evidence indicates that genetic mutations in kinase signaling pathways may modulate treatment responses and clinical outcomes in some cancers 14,15. Recently, we found that KRAS mutation status was a valuable predictor of the tumor response and long-term survival of LARC patients who received preoperative CRT 16. The phosphoinositide-3-kinase (PI3K)/phosphatase and tensinhomolog (PTEN)/v-akt murine thymoma viral oncogene homolog (AKT)/mammalian target of rapamycin (mTOR) signaling pathway participates in tumorigenesis, cancer progression, and drug resistance 17,18. Somatic alterations in the PI3K/PTEN/AKT/mTOR pathway have been implicated as modulators of clinical outcomes in patients with esophageal cancer and lung cancer who underwent CRT 19,20. These studies identified variant single-nucleotide polymorphisms (SNPs) in the PI3K/PTEN/AKT/mTOR pathway 19,20; however, to our knowledge, no studies have reported the relationship between genetic variations in SNPs in the PI3K/PTEN/AKT/mTOR pathway and clinical outcomes in LARC patients who received preoperative CRT. Thus, further study is required to determine the potential clinical value of this approach.
In the current study, we explored the association between genetic variations in 5 genes in this pathway, including AKT1, AKT2, PIK3CA (catalytic subunit of PI3K), PTEN, and FRAP1 (encodes mTOR), and the treatment response and prognosis of LARC patients who received preoperative CRT followed by radical surgery.
Methods
Patient population
This study included consecutive LARC patients treated with preoperative CRT followed by radical surgery between April 2008 and March 2011 at Sun Yat-sen University Cancer Center. All cases were staged based on the 2010 American Joint Committee on Cancer (AJCC) staging system. The inclusion criteria were as follows: (1) histologically confirmed adenocarcinoma; (2) clinical stage of T3-4 or N+ disease; (3) XELOX preoperative chemotherapy (oxaliplatin 130 mg/m2 administered intravenously on day 1 and capecitabine administered orally 1,000 mg/m2 twice daily on days 1-14 for a 3-week cycle); and (4) at least a 3-month postoperative follow-up period. Patients were excluded if they had (1) metastatic disease diagnosed before or during preoperative CRT; (2) less than a 3-month postoperative follow-up period; or (3) another primary malignancy except basal cell carcinoma of the skin. Patient demographics, tumor characteristics, and preoperative treatments were reviewed in detail from patient medical records, and follow-up results were collected from a tracking system. The present study was performed per the ethical standards of the World Medical Association Declaration of Helsinki and approved by the Institutional Research Ethics Committee of Sun Yat-sen University Cancer Center. Informed consent for using pretreatment tissue samples was obtained from the patients.
Treatment and study endpoints
All patients were scheduled to receive a total preoperative irradiation dose of 50 Gy, delivered in fractions of 2.0 Gy daily on 5 consecutive days per week. Concurrent chemotherapy was administered as two cycles of the XELOX regimen. The chemotherapy doses were adjusted by the treatment tolerance. All patients were scheduled for a standard TME within 6 to 8 weeks after completing preoperative radiotherapy. The tumor response to preoperative CRT was assessed using the Dworak pathologic tumor regression grading (TRG) system 21. pCR was defined as the absence of viable tumor cells with only the presence of fibrotic masses or cellular mucin pools in the area occupied by the primary tumor. Briefly, TRG 1 indicates a poor response, and TRG 2-4 indicates a significant response. After radical surgery, all patients were asked to return for a subsequent visit every 3 months for 2 years and then semi-annually for 3 years. Chest/abdominal/pelvic computed tomography (CT) and a colonoscopy were performed annually. Overall survival (OS) was defined as the interval (in months) from the date of surgery until the date of death from any cause or the last follow-up date, while disease-free survival (DFS) was defined as the interval (in months) from the date of surgery to disease recurrence, death or the last follow-up in May 2016.
SNP selection and genotyping
We selected 16 tagging SNPs from 400 bps, flanking and within the gene regions, of five core genes, including AKT1, AKT2, PIK3CA, PTEN and FRAP1 (mTOR), using the tagger algorithm 22. All selected SNPs met the following criteria: minor allele frequency (MAF) ≥ 0.05 among the Chinese population and a tagger algorithm with r2 > 0.8 based on data from the Centre d'Etude du Polymorphisme Humain samples genotyped by the Hap Map Project (www.hapmap.org) 22. Genomic DNA was extracted from peripheral blood lymphocytes that were obtained within one week prior to preoperative CRT using the QIAamp DNA Blood Maxi kit (QIAgen, Valencia, CA, USA) per the manufacturer's protocol. As shown in Supplemental Table 1, 13 SNP assays were selected for amplification with the TaqMan real-time PCR method using the LightCycler® 480 real-time instrument (Roche Diagnostics, Roche Instrument Center AG, Rotkreuz, Switzerland) per the manufacturer's instructions. Due to interference and AT-rich polymorphism nucleotides, SNP assays, including AKT1:rs3803304, AKT1:rs2498804, and AKT2:rs892119, were selected for analysis by sequencing. Sequencing was performed directly on PCR fragments obtained from genomic DNA. DNA amplification was performed as described above, and PCR fragments were purified using the QIAquick PCR purification kit (QIAgen, Valencia, CA, USA). The BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Carlsbad CA, USA) was used for sequencing, and end-point fluorescence was read by a 3730 Genetic Analyzer (Applied Biosystems, Carlsbad CA, USA) per the manufacturer's instructions.
Statistical analysis
To analyze the CRT and pCR responses, crude odds ratios (ORs) along with the corresponding 95% confidence intervals (CIs) for each SNP were calculated using univariate logistic regression analysis, and adjusted ORs were estimated by using multivariate logistic regression, adjusting for age, sex, clinical stage, tumor differentiation, tumor location, cycles of preoperative chemotherapy and time interval from CRT completion to surgery. Crude hazard ratios (HRs) with 95% CIs for 5-year DFS and OS rates were calculated using the univariate Cox proportional hazard model. Adjusted HRs with 95% CIs were estimated with the multivariate Cox model, adjusting for age, sex, clinical stage, tumor differentiation, tumor location, cycles of preoperative chemotherapy and time interval from CRT completion to surgery. Kaplan-Meier analysis and log-rank tests were used to assess differences in DFS and OS. All analyses were performed using IBM SPSS statistics software, version 21.0 (IBM Corp., Armonk, NY, USA). Two-sided statistical tests were used, and a P value < 0.05 was considered significant.
Results
Patient characteristics
A total of 97 eligible LARC patients who received preoperative CRT and radical surgery were included in this study for further analysis (Table 1). Among the patients, 65 (67.0%) were male and 32 (33.0%) were female, with a median age of 58 years (range, 15-75 years). Fifty patients (36.3%) were diagnosed with stage II disease, and 47 (73.7%) were diagnosed with stage III disease. Ninety-one patients (93.8%) received a planned two-cycle XELOX regimen for preoperative CRT. The median interval from completion of CRT to surgery was 6.4 weeks (range 4.3-11.4 weeks). All patients underwent radical surgery. Fifty-two (53.6%) received an anterior resection, 43 (44.3%) received an abdominal perineal resection, and 2 (2.1%) received the Hartmann procedure. After preoperative CRT, 14 patients (14.4%) obtained pCR. The median follow-up time was 65.7 months (range, 3.0 - 98.0 months). During the follow-up period, 28 patients (28.9%) experienced distant metastasis, and 2 (2.1%) experienced local recurrence. Finally, 22 patients (22.7%) died of tumor progression. The 5-year DFS and OS rates were 70.8% and 77.0% for the entire study population, respectively.
Table 1.
Clinical characteristics of locally advanced rectal cancer patients
| Characteristics | Number of patients (%) |
|---|---|
| Age, years | |
| ≤60 | 61(62.9) |
| >60 | 36(37.1) |
| Gender | |
| Male | 65(67.0) |
| Female | 32(33.0) |
| Tumor location, cm | |
| ≤5 | 60(61.9) |
| >5 | 37(38.1) |
| Clinical stage | |
| II | 50(51.5) |
| III | 47(48.5) |
| Tumor differentiation | |
| Well | 20(20.6) |
| Moderate | 69(71.1) |
| Poor | 8(8.2) |
| TRGa | |
| 1 (poor or no response) | 30(30.9) |
| 2 (moderate response) | 25(25.8) |
| 3 (significant response) | 28(28.9) |
| 4 (complete response) | 14(14.4) |
| Pre-CRT CEA level, ng/mL | |
| ≤5 | 50(51.5) |
| >5 | 47(48.5) |
| Pre-CRT CA199 level, U/mL | |
| ≤35 | 82(84.5) |
| >35 | 15(15.5) |
| Cycles of preoperative XELOXb | |
| 1 | 4(4.1) |
| 2 | 91(93.8) |
| 3 | 2(2.1) |
| Interval from completion of CRT to surgery, week | |
| ≤6 | 33(34.0) |
| >6 | 64(66.0) |
| Surgical procedure | |
| Anterior resection | 52(53.6) |
| Abdominal perineal resection | 43(44.3) |
| Hartmann procedure | 2(2.1) |
| Postoperative recurrence | |
| Total | 30(30.9) |
| Local recurrence | 2(2.1) |
| Liver metastasis | 1(1.0) |
| Lung metastasis | 6(6.2) |
| Brain metastasis | 2(2.1) |
| Bone metastasis | 2(2.1) |
| Multiple metastasis | 6(6.2) |
| Unknown site | 11(11.3) |
Abbreviations: CRT: chemoradiotherapy, TRG: tumor regression grade, CEA: carcinoembryonic antigen.
Footnote:
a Tumor regression was assessed by the Dworak TRG system.
b XELOX chemotherapy was administrated as follows: oxaliplatin 130 mg/m2 administered intravenously on day 1 and capecitabine administered orally 1,000 mg/m2 twice daily on days 1-14 for a 3-week cycle.
Associations between SNPs and tumor response to CRT
The SNP analysis results revealed that AKT1:rs3803304, AKT2:rs892119, PIK3CA:rs7640662 and PTEN:rs12357281 were detected without SNP mutations in all patients. We analyzed the 12 SNPs for associations with tumor response to CRT. Only PTEN:rs12569998 was significantly associated with the tumor response (Table 2). Patients with heterozygous PTEN:rs12569998 responded better to CRT than those with a wild-type genotype (crude OR = 2.625; 95% CI, 1.034-6.662, P = 0.042). This association remained significant after adjusting for age, sex, clinical stage, tumor differentiation, tumor location, cycles of preoperative chemotherapy and interval from completion of CRT to surgery (adjusted OR = 2.825; 95% CI, 1.020-7.820, P = 0.046). Patients carrying at least one variant allele presented a better tumor response to CRT than those with a wild-type genotype (adjusted OR = 2.909, 95% CI, 1.127-7.505, P = 0.027). However, no SNPs were significantly associated with pCR (Table 3).
Table 2.
PI3K/PTEN/AKT/mTOR pathway genotypes and preoperative chemoradiotherapy responses
| SNP genotype | TRG1 (n=30) | TRG2-4 (n=67) | Response rate (%) | Crude OR (95%CI) | P value | Adjusted ORa (95%CI) | P value |
|---|---|---|---|---|---|---|---|
| AKT1:rs1130214 | |||||||
| G/G | 25 | 52 | 67.5 | 1(reference) | 1(reference) | ||
| G/C | 5 | 13 | 72.2 | 1.250(0.401-3.894) | 0.700 | 1.102(0.340-3.576) | 0.871 |
| C/C | 0 | 2 | 100.0 | - | - | ||
| G/C+C/C | 5 | 15 | 75.0 | 1.442(0.471-4.416) | 0.606 | 1.309(0.413-4.147) | 0.647 |
| AKT1:rs2494738 | |||||||
| G/G | 7 | 12 | 63.2 | 1(reference) | 1(reference) | ||
| G/A | 13 | 33 | 71.7 | 1.481(0.478-4.592) | 0.497 | 1.575(0.452-5.480) | 0.475 |
| A/A | 10 | 22 | 68.8 | 1.283(0.389-4.239) | 0.682 | 1.407(0.401-4.935) | 0.594 |
| G/A+A/A | 23 | 55 | 70.5 | 1.395(0.487-3.992) | 1.395 | 1.491(0.478-4.655) | 0.492 |
| AKT1:rs2498804 | |||||||
| T/T | 13 | 33 | 71.7 | 1(reference) | 1(reference) | ||
| G/T | 13 | 24 | 64.9 | 0.727(0.287-1.846) | 0.503 | 0.700(0.263-1.861) | 0.474 |
| G/G | 4 | 10 | 71.4 | 0.985(0.262-3.706) | 0.982 | 1.019(0.254-4.083) | 0.979 |
| G/T+G/G | 17 | 34 | 66.7 | 0.788(0.331-1.874) | 0.590 | 0.775(0.314-1.912) | 0.580 |
| AKT2:rs8100018 | |||||||
| G/G | 19 | 30 | 61.2 | 1(reference) | 1(reference) | ||
| G/C | 9 | 33 | 78.6 | 2.322(0.912-5.913) | 0.077 | 2.106(0.785-5.655) | 0.139 |
| C/C | 2 | 4 | 66.7 | 1.267(0.211-7.602) | 0.796 | 0.810(0.109-6.002) | 0.836 |
| G/C+C/C | 11 | 37 | 77.1 | 2.130(0.879-5.162) | 0.094 | 1.893(0.736-4.872) | 0.186 |
| FRAP1:rs2295080 | |||||||
| T/T | 23 | 44 | 65.7 | 1(reference) | 1(reference) | ||
| G/T | 6 | 19 | 76.0 | 1.655(0.581-4.717) | 0.346 | 1.927(0.650-5.711) | 0.237 |
| G/G | 1 | 4 | 80.0 | 2.091(0.221-19.810) | 0.520 | 2.370(0.242-23.179) | 0.458 |
| G/T+G/G | 7 | 23 | 76.7 | 1.718(0.641-4.599) | 0.282 | 1.992(0.718-5.530) | 0.186 |
| FRAP1:rs11121704 | |||||||
| T/T | 28 | 59 | 67.8 | 1(reference) | 1(reference) | ||
| T/C | 2 | 8 | 80.0 | 1.898(0.378-9.530) | 0.436 | 2.246(0.426-11.856) | 0.340 |
| PIK3CA:rs2699887 | |||||||
| G/G | 26 | 61 | 70.1 | 1(reference) | 1(reference) | ||
| G/A | 4 | 6 | 60.0 | 0.639(0.166-2.456) | 0.515 | 0.655(0.158-2.714) | 0.560 |
| PIK3CA:rs6443624 | |||||||
| C/C | 22 | 54 | 71.1 | 1(reference) | 1(reference) | ||
| A/C | 7 | 12 | 63.2 | 0.698(0.243-2.007) | 0.505 | 0.642(0.206-2.000) | 0.642 |
| A/A | 1 | 1 | 50.0 | 0.407(0.024-6.806) | 0.532 | 0.350(0.012-10.332) | 0.543 |
| A/C+A/A | 8 | 13 | 61.9 | 0.662(0.241-1.819) | 0.424 | 0.611(0.204-1.834) | 0.380 |
| PIK3CA:rs7621329 | |||||||
| C/C | 22 | 54 | 71.1 | 1(reference) | 1(reference) | ||
| C/T | 7 | 12 | 63.2 | 0.698(0.243-2.007) | 0.505 | 0.642(0.206-2.000) | 0.642 |
| T/T | 1 | 1 | 50.0 | 0.407(0.024-6.806) | 0.532 | 0.350(0.012-10.332) | 0.543 |
| C/T+T/T | 8 | 13 | 61.9 | 0.662(0.241-1.819) | 0.424 | 0.611(0.204-1.834) | 0.380 |
| PIK3CA:rs7651265 | |||||||
| A/A | 22 | 53 | 70.7 | 1(reference) | 1(reference) | ||
| G/A | 8 | 13 | 61.9 | 0.675(0.245-1.854) | 0.445 | 0.596(0.197-1.809) | 0.361 |
| G/G | 0 | 1 | 100.0 | - | - | ||
| G/A+G/G | 8 | 14 | 63.6 | 0.726(0.267-1.976) | 0.531 | 0.692(0.234-2.051) | 0.507 |
| PTEN:rs2299939 | |||||||
| A/A | 18 | 51 | 73.9 | 1(reference) | 1(reference) | ||
| A/C | 12 | 16 | 57.1 | 0.471(0.187-1.182) | 0.109 | 0.610(0.082-4.540) | 0.629 |
| PTEN:rs12569998 | |||||||
| T/T | 18 | 24 | 57.1 | 1(reference) | 1(reference) | ||
| G/T | 10 | 35 | 77.8 | 2.625(1.034-6.662) | 0.042 | 2.825(1.020-7.820) | 0.046 |
| G/G | 2 | 8 | 80.0 | 3.000(0.567-15.867) | 0.196 | 3.268(0.541-19.738) | 0.197 |
| G/T+G/G | 12 | 43 | 78.2 | 2.687(1.109-6.511) | 0.029 | 2.909(1.127-7.505) | 0.027 |
Abbreviations: SNP: single-nucleotide polymorphism, TRG: tumor regression grade, OR: odds ratio, CI: confidence interval.
Footnote:
aAdjusted for age, sex, clinical stage, tumor differentiation, tumor location, cycles of preoperative chemotherapy and interval from chemoradiotherapy completion to surgery
Table 3.
PI3K/PTEN/AKT/mTOR pathway genotypes and pathological complete responses to preoperative chemoradiotherapy
| SNP genotype | TRG1-3 (non-pCR, n=83) | TRG4 (pCR, n=14) | pCR rate (%) | Crude OR (95%CI) | P value | Adjusted ORa (95%CI) | P value |
|---|---|---|---|---|---|---|---|
| AKT1:rs1130214 | |||||||
| G/G | 65 | 12 | 15.6 | 1(reference) | 1(reference) | ||
| G/C | 16 | 2 | 11.1 | 0.677(0.138-3.333) | 0.632 | 0.745(0.142-3.914) | 0.728 |
| C/C | 2 | 0 | 0.0 | - | - | ||
| G/C+C/C | 18 | 2 | 10.0 | 0.602(0.123-2.938) | 0.530 | 0.588(0.113-3.070) | 0.588 |
| AKT1:rs2494738 | |||||||
| G/G | 15 | 4 | 21.1 | 1(reference) | 1(reference) | ||
| G/A | 42 | 4 | 8.7 | 0.357(0.790-1.610) | 0.180 | 0.348(0.065-1.868) | 0.218 |
| A/A | 26 | 6 | 18.8 | 0.865(0.210-3.565) | 0.841 | 0.987(0.216-4.502) | 0.987 |
| G/A+A/A | 68 | 10 | 12.8 | 0.551(0.152-1.998) | 0.365 | 0.618(0.150-2.549) | 0.506 |
| AKT1:rs2498804 | |||||||
| T/T | 38 | 8 | 17.4 | 1(reference) | 1(reference) | ||
| G/T | 34 | 3 | 8.1 | 0.419(0.103-1.709) | 0.225 | 0.381(0.086-1.695) | 0.205 |
| G/G | 11 | 3 | 21.4 | 1.295(0.293-5.730) | 0.733 | 0.980(0.198-4.844) | 0.981 |
| G/T+G/G | 45 | 6 | 11.8 | 0.633(0.202-1.987) | 0.434 | 0.551(0.164-1.855) | 0.336 |
| AKT2:rs8100018 | |||||||
| G/G | 41 | 8 | 16.3 | 1(reference) | 1(reference) | ||
| G/C | 36 | 6 | 14.3 | 0.854(0.271-2.696) | 0.788 | 1.044(0.302-3.607) | 0.946 |
| C/C | 6 | 0 | 0.0 | - | - | ||
| G/C+C/C | 42 | 6 | 14.3 | 0.732(0.234-2.295) | 0.593 | 0.956(0.276-3.318) | 0.944 |
| FRAP1:rs2295080 | |||||||
| T/T | 56 | 11 | 16.4 | 1(reference) | 1(reference) | ||
| G/T | 23 | 2 | 8.0 | 0.443(0.091-2.155) | 0.313 | 0.343(0.066-1.790) | 0.204 |
| G/G | 4 | 1 | 20.0 | 1.273(0.130-12.501) | 0.836 | 1.303(0.112-15.177) | 0.833 |
| G/T+G/G | 27 | 3 | 10.0 | 0.566(0.146-2.197) | 0.410 | 0.457(0.109-1.909) | 0.283 |
| FRAP1:rs11121704 | |||||||
| T/T | 74 | 13 | 14.9 | 1(reference) | 1(reference) | ||
| T/C | 9 | 1 | 10.0 | 0.632(0.074-5.421) | 0.676 | 0.532(0.057-4.939) | 0.579 |
| PIK3CA:rs2699887 | |||||||
| G/G | 74 | 13 | 14.9 | 1(reference) | 1(reference) | ||
| G/A | 9 | 1 | 10.0 | 0.672(0.074-5.421) | 0.676 | 0.663(0.070-6.292) | 0.720 |
| PIK3CA:rs6443624 | |||||||
| C/C | 65 | 11 | 14.5 | 1(reference) | 1(reference) | ||
| A/C | 16 | 3 | 15.8 | 1.108(0.276-4.443) | 0.885 | 0.949(0.212-4.244) | 0.945 |
| A/A | 2 | 0 | 0.0 | - | - | ||
| A/C+A/A | 18 | 3 | 14.3 | 0.985(0.248-3.912) | 0.983 | 0.856(0.195-3.756) | 0.837 |
| PIK3CA:rs7621329 | |||||||
| C/C | 65 | 11 | 14.5 | 1(reference) | 1(reference) | ||
| C/T | 16 | 3 | 15.8 | 1.108(0.276-4.443) | 0.885 | 0.949(0.212-4.244) | 0.949 |
| T/T | 2 | 0 | 0.0 | - | - | ||
| C/T+T/T | 18 | 3 | 14.3 | 0.985(0.248-3.912) | 0.983 | 0.856(0.195-3.756) | 0.837 |
| PIK3CA:rs7651265 | |||||||
| A/A | 64 | 11 | 14.7 | 1(reference) | 1(reference) | ||
| G/A | 18 | 3 | 14.3 | 0.970(0.244-3.853) | 0.965 | 0.833(0.187-3.713) | 0.810 |
| G/G | 1 | 0 | 0.0 | - | - | ||
| G/A+G/G | 19 | 3 | 13.6 | 0.919(0.232-3.635) | 0.904 | 0.769(0.175-3.373) | 0.728 |
| PTEN:rs2299939 | |||||||
| A/A | 58 | 11 | 15.9 | 1(reference) | 1(reference) | ||
| A/C | 25 | 3 | 10.7 | 0.633(0.162-2.465) | 0.509 | 0.560(0.129-2.426) | 0.438 |
| PTEN:rs12569998 | |||||||
| T/T | 38 | 4 | 9.5 | 1(reference) | 1(reference) | ||
| G/T | 37 | 8 | 17.8 | 2.054(0.570-7.408) | 0.271 | 2.136(0.546-8.357) | 0.276 |
| G/G | 8 | 2 | 20.0 | 2.375(0.369-15.268) | 0.362 | 2.703(0.346-21.083) | 0.343 |
| G/T+G/G | 45 | 10 | 18.2 | 2.111(0.613-7.276) | 0.237 | 2.242(0.614-8.185) | 0.222 |
Abbreviations: SNP: single-nucleotide polymorphism, TRG: tumor regression grade, pCR: pathological complete response, OR: odds ratio, CI: confidence interval.
Footnote:
aAdjusted for age, sex, clinical stage, tumor differentiation, tumor location, cycles of preoperative chemotherapy and interval from chemoradiotherapy completion to surgery.
Associations between SNPs and DFS and OS
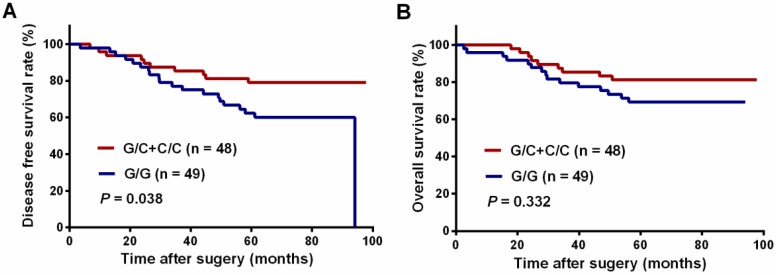
We next analyzed the association between SNPs and long-term survival. AKT2:rs8100018 was identified as the only SNP associated with 5-year DFS among the 12 SNPs (Table 4). Patients with heterozygous AKT2:rs8100018 showed a decreased recurrence risk and a better 5-year DFS than those with a wild-type genotype (crude HR = 0.358; 95% CI, 0.151-0.848, P = 0.020). This association remained significant after adjusting for age, sex, clinical stage, tumor differentiation, tumor location, cycles of preoperative chemotherapy and interval from completion of CRT to surgery (adjusted HR = 0.329; 95% CI, 0.135-0.802, P = 0.014). Furthermore, patients carrying at least one variant allele also showed a significantly reduced recurrence risk (adjusted HR = 0.414; 95% CI, 0.187-0.914, P = 0.029). As shown in Figure 1A, patients carrying at least one variant allele in AKT2:rs8100018 presented a higher 5-year DFS rate than those with a wild-type genotype (79.2% vs. 62.3%, P = 0.038). Although patients carrying at least one variant allele in AKT2:rs8100018 had a higher 5-year OS rate than those with a wild-type genotype, no significant difference was found between the groups (81.3% vs. 72.7%, P = 0.332, Figure 1B). In addition, there was no significant association between any genotypes for the 12 SNPs and 5-year OS (Table 5).
Table 4.
PI3K/PTEN/AKT/mTOR pathway genotypes and disease free survival after preoperative chemoradiotherapy and surgery
| SNP genotype | No Recurrence | Recurrence | 5-year DFS rate (%) | Crude HR (95%CI) | P value | Adjusted HRa (95%CI) | P value |
|---|---|---|---|---|---|---|---|
| AKT1:rs1130214 | |||||||
| G/G | 53 | 24 | 71.0 | 1(reference) | 1(reference) | ||
| G/C | 12 | 6 | 66.7 | 1.040(0.424-2.556) | 0.931 | 1.064(0.429-2.640) | 0.894 |
| C/C | 2 | 0 | - | - | - | ||
| G/C+C/C | 14 | 6 | 70.0 | 0.923(0.376-2.268) | 0.862 | 0.938(0.378-2.330) | 0.891 |
| AKT1:rs2494738 | |||||||
| G/G | 14 | 5 | 73.7 | 1(reference) | 1(reference) | ||
| G/A | 32 | 14 | 69.4 | 1.120(0.403-3.116) | 0.828 | 1.554(0.529-4.566) | 0.423 |
| A/A | 21 | 11 | 71.0 | 1.349(0.466-3.902) | 0.581 | 1.506(0.515-4.404) | 0.455 |
| G/A+A/A | 53 | 25 | 70.1 | 1.209(0.461-3.170) | 0.699 | 1.529(0.568-4.116) | 0.401 |
| AKT1:rs2498804 | |||||||
| T/T | 32 | 14 | 71.1 | 1(reference) | 1(reference) | ||
| G/T | 24 | 13 | 67.4 | 1.062(0.499-2.261) | 0.876 | 1.164(0.529-2.557) | 0.706 |
| G/G | 11 | 3 | 78.6 | 0.649(0.186-2.264) | 0.498 | 0.596(0.169-2.105) | 0.421 |
| G/T+G/G | 35 | 16 | 70.4 | 0.949(0.463-1.946) | 0.887 | 0.976(0.468-2.034) | 0.949 |
| AKT2:rs8100018 | |||||||
| G/G | 29 | 20 | 62.3 | 1(reference) | 1(reference) | ||
| G/C | 35 | 7 | 83.3 | 0.358(0.151-0.848) | 0.020 | 0.329(0.135-0.802) | 0.014 |
| C/C | 3 | 3 | 50.0 | 1.273(0.377-4.300) | 0.697 | 1.165(0.298-4.553) | 0.827 |
| G/C+C/C | 38 | 10 | 79.2 | 0.457(0.214-0.977) | 0.043 | 0.414(0.187-0.914) | 0.029 |
| FRAP1:rs2295080 | |||||||
| T/T | 46 | 21 | 71.5 | 1(reference) | 1(reference) | ||
| G/T | 18 | 7 | 70.8 | 0.913(0.386-2.160) | 0.836 | 0.909(0.376-2.197) | 0.863 |
| G/G | 3 | 2 | 60.0 | 1.298(0.303-5.555) | 0.725 | 1.289(0.296-5.607) | 0.735 |
| G/T+G/G | 21 | 9 | 69.0 | 0.978(0.445-2.148) | 0.995 | 0.973(0.433-2.186) | 0.947 |
| FRAP1:rs11121704 | |||||||
| T/T | 58 | 29 | 68.5 | 1(reference) | 1(reference) | ||
| T/C | 9 | 1 | 90.0 | 0.260(0.035-1.911) | 0.186 | 0.275(0.370-2.051) | 0.208 |
| PIK3CA:rs2699887 | |||||||
| G/G | 60 | 27 | 70.9 | 1(reference) | 1(reference) | ||
| G/A | 7 | 3 | 70.0 | 0.871(0.264-2.879) | 0.821 | 1.095(0.316-3.801) | 0.886 |
| PIK3CA:rs6443624 | |||||||
| C/C | 54 | 22 | 73.6 | 1(reference) | 1(reference) | ||
| A/C | 11 | 8 | 57.9 | 1.645(0.728-3.716) | 0.232 | 2.314(0.949-5.644) | 0.065 |
| A/A | 2 | 0 | - | - | - | ||
| A/C+A/A | 13 | 8 | 60.0 | 1.541(0.682-3.482) | 0.298 | 2.245(0.918-5.492) | 0.076 |
| PIK3CA:rs7621329 | |||||||
| C/C | 54 | 22 | 73.6 | 1(reference) | 1(reference) | ||
| C/T | 11 | 8 | 57.9 | 1.645(0.728-3.716) | 0.232 | 2.314(0.949-5.644) | 0.065 |
| T/T | 2 | 0 | - | - | - | ||
| C/T+T/T | 13 | 8 | 60.0 | 1.541(0.682-3.482) | 0.298 | 2.245(0.918-5.492) | 0.076 |
| PIK3CA:rs7651265 | |||||||
| A/A | 53 | 22 | 73.2 | 1(reference) | 1(reference) | ||
| G/A | 13 | 8 | 61.9 | 1.428(0.632-3.226) | 0.391 | 2.012(0.823-4.922) | 0.126 |
| G/G | 1 | 0 | - | - | - | ||
| G/A+G/G | 14 | 8 | 61.9 | 1.428(0.632-3.226) | 0.391 | 2.012(0.823-4.922) | 0.126 |
| PTEN:rs2299939 | |||||||
| A/A | 48 | 21 | 72.4 | 1(reference) | 1(reference) | ||
| A/C | 19 | 9 | 66.7 | 1.26(0.574-2.768) | 0.565 | 1.327(0.583-3.020) | 0.500 |
| PTEN:rs12569998 | |||||||
| T/T | 29 | 13 | 70.7 | 1(reference) | 1(reference) | ||
| G/T | 29 | 16 | 66.3 | 1.037(0.495-2.171) | 0.924 | 1.230(0.568-2.664) | 0.600 |
| G/G | 9 | 1 | 90.0 | 0.216(0.270-1.701) | 0.145 | 0.163(0.221-1.844) | 0.163 |
| G/T+G/G | 38 | 17 | 70.7 | 0.866(0.417-1.801) | 0.701 | 0.993(0.466-2.119) | 0.986 |
Abbreviations: SNP: single-nucleotide polymorphism, DFS: disease-free survival, HR: hazard ratio, CI: confidence interval.
Footnote:
aAdjusted for age, sex, clinical stage, tumor differentiation, tumor location, cycles of preoperative chemotherapy and interval from chemoradiotherapy completion to surgery.
Figure 1.
Kaplan-Meier curves of patients with locally advanced rectal cancer grouped by their AKT2:rs8100018 genotype. (A) The disease-free survival (DFS) and (B) overall survival (OS) of all patients.
Table 5.
PI3K/PTEN/AKT/mTOR pathway genotypes and overall survival after preoperative chemoradiotherapy and surgery
| SNP genotype | Alive | Dead | 5-year OS rate (%) | Crude HR (95%CI) | P value | Adjusted HRa (95%CI) | P value |
|---|---|---|---|---|---|---|---|
| AKT1:rs1130214 | |||||||
| G/G | 58 | 19 | 74.9 | 1(reference) | 1(reference) | ||
| G/C | 15 | 3 | 83.3 | 0.628(0.186-2.123) | 0.454 | 0.632(0.186-2.153) | 0.463 |
| C/C | 2 | 0 | - | - | - | ||
| G/C+C/C | 17 | 3 | 85.0 | 0.561(0.166-1.895) | 0.352 | 0.555(0.163-1.889) | 0.347 |
| AKT1:rs2494738 | |||||||
| G/G | 16 | 3 | 84.2 | 1(reference) | 1(reference) | ||
| G/A | 35 | 11 | 76.0 | 1.586(0.443-5.687) | 0.479 | 1.992(0.523-7.594) | 0.313 |
| A/A | 24 | 8 | 74.2 | 1.768(0.469-6.665) | 0.400 | 1.967(0.517-7.485) | 0.321 |
| G/A+A/A | 59 | 19 | 75.3 | 1.658(0.491-5.603) | 0.416 | 1.979(0.568-6.897) | 0.284 |
| AKT1:rs2498804 | |||||||
| T/T | 35 | 11 | 75.6 | 1(reference) | 1(reference) | ||
| G/T | 27 | 10 | 72.9 | 1.059(0.449-2.494) | 0.896 | 1.171(0.480-2.856) | 0.729 |
| G/G | 13 | 1 | 92.9 | 0.263(0.034-2.039) | 0.201 | 0.229(0.0229-1.807) | 0.162 |
| G/T+G/G | 40 | 11 | 78.4 | 0.830(0.360-1.915) | 0.663 | 0.840(0.358-1.967) | 0.687 |
| AKT2:rs8100018 | |||||||
| G/G | 36 | 13 | 72.7 | 1(reference) | 1(reference) | ||
| G/C | 35 | 7 | 83.3 | 0.581(0.232-1.455) | 0.246 | 0.544(0.210-1.410) | 0.210 |
| C/C | 4 | 2 | 66.7 | 1.243(0.280-5.510) | 0.775 | 1.229(0.235-6.416) | 0.807 |
| G/C+C/C | 39 | 9 | 81.3 | 0.659(0.282-1.541) | 0.336 | 0.615(0.253-1.492) | 0.282 |
| FRAP1:rs2295080 | |||||||
| T/T | 50 | 17 | 74.6 | 1(reference) | 1(reference) | ||
| G/T | 22 | 3 | 87.3 | 0.452(0.132-0.142) | 0.205 | 0.454(0.131-1.566) | 0.211 |
| G/G | 3 | 2 | 60.0 | 1.630(0.376-7.058) | 0.514 | 1.690(0.384-7.444) | 0.488 |
| G/T+G/G | 25 | 5 | 82.5 | 0.636(0.234-1.723) | 0.373 | 0.640(0.233-1.760) | 0.388 |
| FRAP1:rs11121704 | |||||||
| T/T | 65 | 22 | 74.4 | 1(reference) | 1(reference) | ||
| T/C | 10 | 0 | - | 0.041(0.001-12.434) | 0.274 | - | |
| PIK3CA:rs2699887 | |||||||
| G/G | 67 | 20 | 76.7 | 1(reference) | 1(reference) | ||
| G/A | 8 | 2 | 80.0 | 0.784(0.183-3.354) | 0.742 | 0.902(0.202-4.039) | 0.902 |
| PIK3CA:rs6443624 | |||||||
| C/C | 61 | 15 | 80.2 | 1(reference) | 1(reference) | ||
| A/C | 12 | 7 | 63.2 | 1.973(0.804-4.840) | 0.138 | 2.391(0.916-6.237) | 0.075 |
| A/A | 2 | 0 | - | - | - | ||
| A/C+A/A | 14 | 7 | 65.0 | 1.857(0.757-4.555) | 0.177 | 2.326(0.889-6.085) | 0.085 |
| PIK3CA:rs7621329 | |||||||
| C/C | 61 | 15 | 80.2 | 1(reference) | 1(reference) | ||
| C/T | 12 | 7 | 63.2 | 1.973(0.804-4.840) | 0.138 | 2.391(0.916-6.237) | 0.075 |
| T/T | 2 | 0 | - | - | - | ||
| C/T+T/T | 14 | 7 | 65.0 | 1.857(0.757-4.555) | 0.177 | 2.326(0.889-6.085) | 0.085 |
| PIK3CA:rs7651265 | |||||||
| A/A | 60 | 15 | 80.0 | 1(reference) | 1(reference) | ||
| G/A | 14 | 7 | 66.7 | 1.728(0.704-4.238) | 1.728 | 2.126(0.812-5.563) | 0.124 |
| G/G | 1 | 0 | - | - | - | ||
| G/A+G/G | 15 | 7 | 66.7 | 1.728(0.704-4.238) | 1.728 | 2.126(0.812-5.563) | 0.124 |
| PTEN:rs2299939 | |||||||
| A/A | 54 | 15 | 78.3 | 1(reference) | 1(reference) | ||
| A/C | 21 | 7 | 74.1 | 1.277(0.521-3.134) | 0.593 | 1.330(0.522-3.386) | 0.550 |
| PTEN:rs12569998 | |||||||
| T/T | 32 | 10 | 75.6 | 1(reference) | 1(reference) | ||
| G/T | 34 | 11 | 75.4 | 0.998(0.424-2.350) | 0.996 | 1.141(0.463-2.811) | 0.774 |
| G/G | 9 | 1 | 90.0 | 0.358(0.046-2.796) | 0.327 | 0.330(0.040-2.711) | 0.302 |
| G/T+G/G | 43 | 12 | 78.0 | 0.869(0.375-2.011) | 0.742 | 0.936(0.392-2.231) | 0.881 |
Abbreviations: SNP: single-nucleotide polymorphism, OS: overall survival, HR: hazard ratio, CI: confidence interval.
Footnote:
aAdjusted for age, sex, clinical stage, tumor differentiation, tumor location, cycles of preoperative chemotherapy and interval from chemoradiotherapy completion to surgery.
Discussion
The PI3K/PTEN/AKT/mTOR pathway is involved in tumor proliferation, differentiation, cell survival, trafficking, and glucose homeostasis. Therefore, studying the PI3K/PTEN/AKT/mTOR pathway in human cancer may provide potential drug targets for therapeutic intervention 23-25. Currently, there is a lack of suitable biomarkers for assessing this signaling pathway in LARC patients receiving preoperative CRT and radical surgery. In the present study, we determined the regulation status of the PI3K/PTEN/AKT/mTOR pathway by applying tagging SNPs to detect the genetic variations of PI3K, PTEN, AKT1, AKT2, and mTOR in LARC patients. We found that SNPs in PTEN:rs12569998 were associated with tumor response, and AKT2:rs8100018 was associated with 5-year DFS. To the best of our knowledge, this is the first study revealing this clinical implication of SNPs in the PI3K/PTEN/AKT/mTOR pathway in LARC patients treated with preoperative CRT and radical surgery.
It is worth noting that the activation status of the PI3K/PTEN/AKT/mTOR pathway can influence the tumor response to CRT. Evidence has shown that this pathway is associated with the response to platinum-based chemotherapy treatment in several cancers 26-28. For rectal cancer patients, oxaliplatin is the most common platinum-containing chemotherapeutic agent. We found that individuals carrying heterozygous PTEN:rs12569998 responded better to an oxaliplatin-based preoperative chemotherapeutic regimen. As a tumor suppressor, PTEN dephosphorylates PI3K at the 3-phosphate site and negatively regulates the Akt signal pathway to inhibit cell proliferation and facilitate apoptosis 29. In contrast, aberrantly altering PTEN promotes cell cycle progression by disrupting genetic transmission and inactivating cell cycle checkpoints 30. Therefore, tumor cells with variant PTEN expression may be more sensitive to oxaliplatin-based chemotherapy than those with more cells in the G(0)/G(1) phases and fewer cells in the G(2)/M phases 31. Moreover, PI3K/PTEN/AKT/mTOR pathway activity leads to different tumor cell chemotherapy and radiotherapy responses 32,33. The cellular response to irradiation is driven by direct DNA damage in the targeted cells. Further characterization revealed a PI3K/AKT-independent mechanism of actin remodeling that is responsible for PTEN function in controlling the DNA damage-inducible cell size checkpoint 34. Recent findings have shown that PTEN is also critical in DNA damage repair and the DNA damage response (DDR) 35. In particular, PTEN loss decreases Chk1 and p53 activity by regulating their protein levels and promoting genomic instability in tumor cells 36,37. Based on these results, we considered that the variant PTEN:rs12569998 may represent PTEN dysfunction, thereby impairing the DDR. This may indicate that rectal tumors with heterozygous PTEN:rs12569998 may be sensitive to radiation and thus respond better to preoperative CRT.
A significant association was observed between AKT2:rs8100018 and DFS in our study. Playing a central role in the PI3K pathway, genetic alterations in AKT influence the downstream-activated effect in oncogenesis and thus highlight their distinct functional roles in cancer development and progression 38. Several studies have reported that activated AKT2 expression positively correlates with aggressiveness and poor prognosis in glioma, hepatocellular, and colorectal tumors 39-41. To our knowledge, data on the associations between common genetic variations in AKT2 and oncologic outcomes are limited in LARC patients. Hildebrandt MA et al. first found that AKT2 SNP rs892119 was significantly associated with an increased risk of recurrence and death 19. This finding may suggest that the functional SNP tagged by AKT2:rs892119 activates AKT2 and increases signaling through the PI3K/PTEN/AKT/mTOR pathway. Pfisterer K et al. revealed that patients with variants of AKT2:rs8100018 had a higher risk of progression after docetaxel and cetuximab and experienced a shorter progression-free survival in squamous cell carcinomas of the head and neck 42. However, in contrast to these results, our study found that patients with AKT2:rs8100018 variants had a lower risk of recurrence after receiving preoperative CRT followed by radical surgery and experienced a longer DFS. This indicates that these functional variants diminish AKT2 activity and decrease signaling through this pathway, thus reducing cell survival signals in rectal tumors. It is also likely that AKT2:rs810001 is not a functional variant, but a surrogate marker, for the underlying genetic variation within that genomic region. In addition, the prognostic impact of AKT2 may be affected by different cancers, treatment strategies, and follow-up schedules. As the variants genotyped in this study were tagging SNPs, we cannot identify the causative SNPs and mechanisms responsible. Thus, additional studies are required to identify the prognostic value of AKT2 variation in an extended cohort of LARC patients.
Several limitations should be addressed in this study. First, this retrospective study included an uncontrolled methodology and a limited number of patients from a single institution. Accordingly, we could not rule out the possibility of false-positive findings. Therefore, the findings need to be validated in a larger cohort of patients. Second, this study only included tagging SNPs of five genes, and other genes related to this pathway warrant investigation, including TSC1, SMAD and PDK1 43-45. This approach may have led to the omission of additional functionally relevant prognostic SNPs. Additionally, complete data on the patients' postoperative treatments were unavailable. Thus, we could not adjust the ORs of HRs with a postoperative treatment factor. Finally, since the variants genotyped in this study were tagging SNPs, we were unable to identify all causative SNPs and the mechanisms responsible. However, this approach can predict tumor responses and determine which patients are at risk for postoperative disease recurrence. Once patients are determined to have unfavorable genotypes, these patients will likely receive no benefit from preoperative CRT, and thus only surgery and more frequent postoperative follow-ups should be recommended.
Conclusion
This is the first study to indicate the association between genetic variations in the PI3K/PTEN/AKT/mTOR pathway and the clinical outcomes of LARC patients treated with preoperative CRT and radical surgery. We found that PTEN:rs12569998 was associated with the tumor response and AKT2:rs8100018 was associated with 5-year DFS after preoperative CRT. The clinical value of tagging SNPs in the PI3K/PTEN/AKT/mTOR pathway requires further investigation in LARC patients.
Supplementary Material
Supplementary table S1.
Acknowledgments
We would like to thank the all colleagues of Department of Colorectal Surgery in Sun Yat-sen University Cancer Center, who have involved with performing the treatment for current study. We also thank Yixin Zhao, Shanshan Sun, and Diefei Liao for offering insightful effort in this study. The authenticity of this article has been validated by uploading the key raw data onto the Research Data.
Deposit public platform (http://www.researchdata.org.cn), with the approval number as RDDB2017000165.
Funding
This work was funded by grants from National Natural Science Foundation of China (No.81772595 and No. 81502459), the Sun Yat-sen University Clinical Research 5010 Program (No.2015024 and No.2013013) and Science and Technology Planning Project of Guangdong Province (No 2016ZC0028).
Authors' contributions
JHP and WJM carried out the data analysis and drafted the manuscript; ZGZ, YKG and ZHL participated in clinical data collection; RXZ, JHP and WJM performed SNP selection and genotyping. RXZ and ZZP supervised the research program and edited the manuscript; RXZ and ZZP had significant roles in the study design and manuscript review. All authors read and approved the final manuscript.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Glimelius B, Tiret E, Cervantes A, Arnold D. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(Suppl 6):i81–8. doi: 10.1093/annonc/mdt240. [DOI] [PubMed] [Google Scholar]
- 4.Benson AR, Venook AP, Bekaii-Saab T, Chan E, Chen YJ, Cooper HS. et al. Rectal Cancer, Version 2.2015. J Natl Compr Canc Netw. 2015;13:719–28. doi: 10.6004/jnccn.2015.0087. [DOI] [PubMed] [Google Scholar]
- 5.Peng J, Lin J, Qiu M, Wu X, Lu Z, Chen G. et al. Clinical factors of post-chemoradiotherapy as valuable indicators for pathological complete response in locally advanced rectal cancer. Clinics (Sao Paulo) 2016;71:449–54. doi: 10.6061/clinics/2016(08)07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao YH, Lin JZ, An X, Luo JL, Cai MY, Cai PQ. et al. Neoadjuvant sandwich treatment with oxaliplatin and capecitabine administered prior to, concurrently with, and following radiation therapy in locally advanced rectal cancer: a prospective phase 2 trial. Int J Radiat Oncol Biol Phys. 2014;90:1153–60. doi: 10.1016/j.ijrobp.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 7.Lin JZ, Zeng ZF, Wu XJ, Wan DS, Chen G, Li LR. et al. Phase II study of pre-operative radiotherapy with capecitabine and oxaliplatin for rectal cancer and carcinoembryonic antigen as a predictor of pathological tumour response. J Int Med Res. 2010;38:645–54. doi: 10.1177/147323001003800227. [DOI] [PubMed] [Google Scholar]
- 8.Park IJ, You YN, Agarwal A, Skibber JM, Rodriguez-Bigas MA, Eng C. et al. Neoadjuvant treatment response as an early response indicator for patients with rectal cancer. J Clin Oncol. 2012;30:1770–6. doi: 10.1200/JCO.2011.39.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodel C, Martus P, Papadoupolos T, Fuzesi L, Klimpfinger M, Fietkau R. et al. Prognostic significance of tumor regression after preoperative chemoradiotherapy for rectal cancer. J Clin Oncol. 2005;23(34):8688–96. doi: 10.1200/JCO.2005.02.1329. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Peng J, Qdaisat A, Li L, Chen G, Lu Z. et al. Severe weight loss during preoperative chemoradiotherapy compromises survival outcome for patients with locally advanced rectal cancer. J Cancer Res Clin Oncol. 2016;142:2551–60. doi: 10.1007/s00432-016-2225-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin JZ, Peng JH, Qdaisat A, Lu ZH, Wu XJ, Chen G. et al. Preoperative chemoradiotherapy creates an opportunity to perform sphincter preserving resection for low-lying locally advanced rectal cancer based on an oncologic outcome study. Oncotarget. 2016;7:57317–26. doi: 10.18632/oncotarget.10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valentini V, van Stiphout RG, Lammering G, Gambacorta MA, Barba MC, Bebenek M. et al. Nomograms for predicting local recurrence, distant metastases, and overall survival for patients with locally advanced rectal cancer on the basis of European randomized clinical trials. J Clin Oncol. 2011;29:3163–72. doi: 10.1200/JCO.2010.33.1595. [DOI] [PubMed] [Google Scholar]
- 13.Zeng WG, Liang JW, Wang Z, Zhang XM, Hu JJ, Hou HR. et al. Clinical parameters predicting pathologic complete response following neoadjuvant chemoradiotherapy for rectal cancer. Chin J Cancer. 2015;34:468–74. doi: 10.1186/s40880-015-0033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakrabarti G. Mutant KRAS associated malic enzyme 1 expression is a predictive marker for radiation therapy response in non-small cell lung cancer. Radiat Oncol. 2015;10:145. doi: 10.1186/s13014-015-0457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang ZC, Fu S, Wang F, Wang HY, Zeng YX, Shao JY. Oncogene mutational profile in nasopharyngeal carcinoma. Onco Targets Ther. 2014;7:457–67. doi: 10.2147/OTT.S58791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng J, Lin J, Qiu M, Zhao Y, Deng Y, Shao J. et al. Oncogene mutation profile predicts tumor regression and survival in locally advanced rectal cancer patients treated with preoperative chemoradiotherapy and radical surgery. Tumour Biol. 2017;39:1393380026. doi: 10.1177/1010428317709638. [DOI] [PubMed] [Google Scholar]
- 17.Costa C, Pereira S, Lima L, Peixoto A, Fernandes E, Neves D. et al. Abnormal Protein Glycosylation and Activated PI3K/Akt/mTOR Pathway: Role in Bladder Cancer Prognosis and Targeted Therapeutics. Plos One. 2015;10:e141253. doi: 10.1371/journal.pone.0141253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Dang Y, Gao J, Li Y, Zou J, Shen L. PI3K/AKT/mTOR pathway is activated after imatinib secondary resistance in gastrointestinal stromal tumors (GISTs) Med Oncol. 2015;32:111. doi: 10.1007/s12032-015-0554-6. [DOI] [PubMed] [Google Scholar]
- 19.Hildebrandt MA, Yang H, Hung MC, Izzo JG, Huang M, Lin J. et al. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. J Clin Oncol. 2009;27:857–71. doi: 10.1200/JCO.2008.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pu X, Hildebrandt MA, Lu C, Lin J, Stewart DJ, Ye Y. et al. PI3K/PTEN/AKT/mTOR pathway genetic variation predicts toxicity and distant progression in lung cancer patients receiving platinum-based chemotherapy. Lung Cancer. 2011;71:82–8. doi: 10.1016/j.lungcan.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dworak O, Keilholz L, Hoffmann A. Pathological features of rectal cancer after preoperative radiochemotherapy. Int J Colorectal Dis. 1997;12:19–23. doi: 10.1007/s003840050072. [DOI] [PubMed] [Google Scholar]
- 22.A haplotype map of the human genome. Nature. 2005; 437:1299-320. [DOI] [PMC free article] [PubMed]
- 23.Lim HJ, Crowe P, Yang JL. Current clinical regulation of PI3K/PTEN/Akt/mTOR signalling in treatment of human cancer. J Cancer Res Clin Oncol. 2015;141:671–89. doi: 10.1007/s00432-014-1803-3. [DOI] [PubMed] [Google Scholar]
- 24.Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4:257–62. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 25.Steelman LS, Chappell WH, Abrams SL, Kempf RC, Long J, Laidler P. et al. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity to therapy-implications for cancer and aging. Aging (Albany NY) 2011;3:192–222. doi: 10.18632/aging.100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu JL, Wang ZW, Hu LM, Yin ZQ, Huang MD, Hu ZB. et al. Genetic variants in the PI3K/PTEN/AKT/mTOR pathway predict platinum-based chemotherapy response of advanced non-small cell lung cancers in a Chinese population. Asian Pac J Cancer Prev. 2012;13:2157–62. doi: 10.7314/apjcp.2012.13.5.2157. [DOI] [PubMed] [Google Scholar]
- 27.Peng DJ, Wang J, Zhou JY, Wu GS. Role of the Akt/mTOR survival pathway in cisplatin resistance in ovarian cancer cells. Biochem Biophys Res Commun. 2010;394:600–5. doi: 10.1016/j.bbrc.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiu P, Dong X, Dong X, Xu Z, Zhu H, Liu F. et al. Secretory clusterin contributes to oxaliplatin resistance by activating Akt pathway in hepatocellular carcinoma. Cancer Sci. 2013;104:375–82. doi: 10.1111/cas.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T. et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 30.Brandmaier A, Hou SQ, Shen WH. Cell Cycle Control by PTEN. J Mol Biol. 2017;429:2265–77. doi: 10.1016/j.jmb.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Qiu M, Tang QL, Liu M, Lang N, Bi F. Establishment and biological characteristics of oxaliplatin-resistant human colon cancer cell lines. Chin J Cancer. 2010;29:661–7. doi: 10.5732/cjc.009.10666. [DOI] [PubMed] [Google Scholar]
- 32.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Franklin RA, Montalto G. et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascade inhibitors: how mutations can result in therapy resistance and how to overcome resistance. Oncotarget. 2012;3:1068–111. doi: 10.18632/oncotarget.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chappell WH, Steelman LS, Long JM, Kempf RC, Abrams SL, Franklin RA. et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget. 2011;2:135–64. doi: 10.18632/oncotarget.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JS, Xu X, Li H, Solomon D, Lane WS, Jin T. et al. Mechanistic analysis of a DNA damage-induced, PTEN-dependent size checkpoint in human cells. Mol Cell Biol. 2011;31:2756–71. doi: 10.1128/MCB.01323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ming M, He YY. PTEN in DNA damage repair. Cancer Lett. 2012;319:125–29. doi: 10.1016/j.canlet.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, Memeo L. et al. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell. 2005;7:193–204. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Freeman DJ, Li AG, Wei G, Li HH, Kertesz N, Lesche R. et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell. 2003;3:117–30. doi: 10.1016/s1535-6108(03)00021-7. [DOI] [PubMed] [Google Scholar]
- 38.Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med. 2014;46:372–83. doi: 10.3109/07853890.2014.912836. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Han L, Zhang A, Wang Y, Yue X, You Y. et al. AKT2 expression is associated with glioma malignant progression and required for cell survival and invasion. Oncol Rep. 2010;24:65–72. doi: 10.3892/or_00000829. [DOI] [PubMed] [Google Scholar]
- 40.Xu X, Sakon M, Nagano H, Hiraoka N, Yamamoto H, Hayashi N. et al. Akt2 expression correlates with prognosis of human hepatocellular carcinoma. Oncol Rep. 2004;11:25–32. [PubMed] [Google Scholar]
- 41.Rychahou PG, Kang J, Gulhati P, Doan HQ, Chen LA, Xiao SY. et al. Akt2 overexpression plays a critical role in the establishment of colorectal cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:20315–20. doi: 10.1073/pnas.0810715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfisterer K, Fusi A, Klinghammer K, Knodler M, Nonnenmacher A, Keilholz U. PI3K/PTEN/AKT/mTOR polymorphisms: association with clinical outcome in patients with head and neck squamous cell carcinoma receiving cetuximab-docetaxel. Head Neck. 2015;37:471–8. doi: 10.1002/hed.23604. [DOI] [PubMed] [Google Scholar]
- 43.Makker A, Goel MM, Mahdi AA. PI3K/PTEN/Akt and TSC/mTOR signaling pathways, ovarian dysfunction, and infertility: an update. J Mol Endocrinol. 2014;53:R103–8. doi: 10.1530/JME-14-0220. [DOI] [PubMed] [Google Scholar]
- 44.Rocher C, Singla DK. SMAD-PI3K-Akt-mTOR pathway mediates BMP-7 polarization of monocytes into M2 macrophages. Plos One. 2013;8:e84009. doi: 10.1371/journal.pone.0084009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hildebrandt MA, Lippman SM, Etzel CJ, Kim E, Lee JJ, Khuri FR. et al. Genetic variants in the PI3K/PTEN/AKT/mTOR pathway predict head and neck cancer patient second primary tumor/recurrence risk and response to retinoid chemoprevention. Clin Cancer Res. 2012;18:3705–13. doi: 10.1158/1078-0432.CCR-11-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table S1.