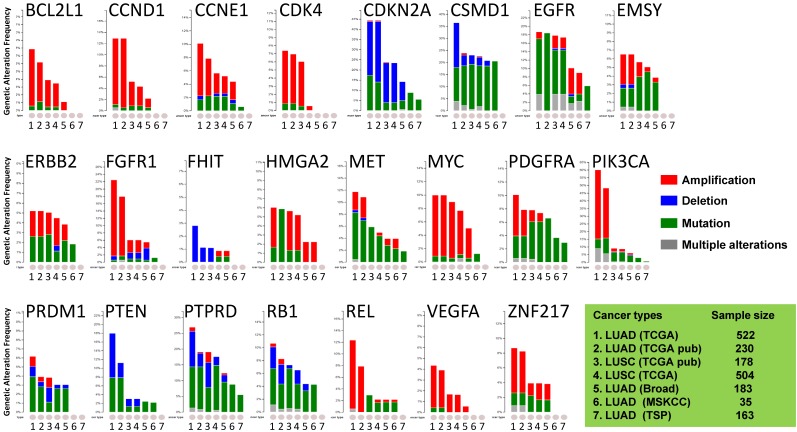
Figure 1.
Public dataset-based analysis of somatic alterations for 23 lung cancer-associate genes associated with lung cancers. A cross-cancer alteration summary for 23 lung cancer-associated genes was prepared using c-Bioportal. Data mining was accomplished with cBioPortal 25, 26 for Cancer Genomics, a data portal (cBioPortal for Cancer Genomics) available at http://www.cbioportal.org, to measure the incidence of conditions that are associated with the alterations in these genes. The database query was based on deregulation (amplification, deletion, and mutation) of these genes. All datasets used were from publically available sources, including Broad 21 (Broad Institute of MIT and Harvard, 183 LUAD samples), MSKCC 22 (Memorial Sloan Kettering Cancer Center, 35 LUAD samples) or various projects, including TCGA 23, 24 (the Cancer Genome Atlas - Cancer Genome, 230 LUAD samples and 178 LUSC samples), TSP 6 (the Tumor Sequencing Project, 163 LUAD samples). "TCGA" means provisional TCGA datasets; "TCGA pub" means TCGA datasets used in the corresponding publications. False discovery rate (FDR) and Bonferroni corrections were applied.