Figure 1.
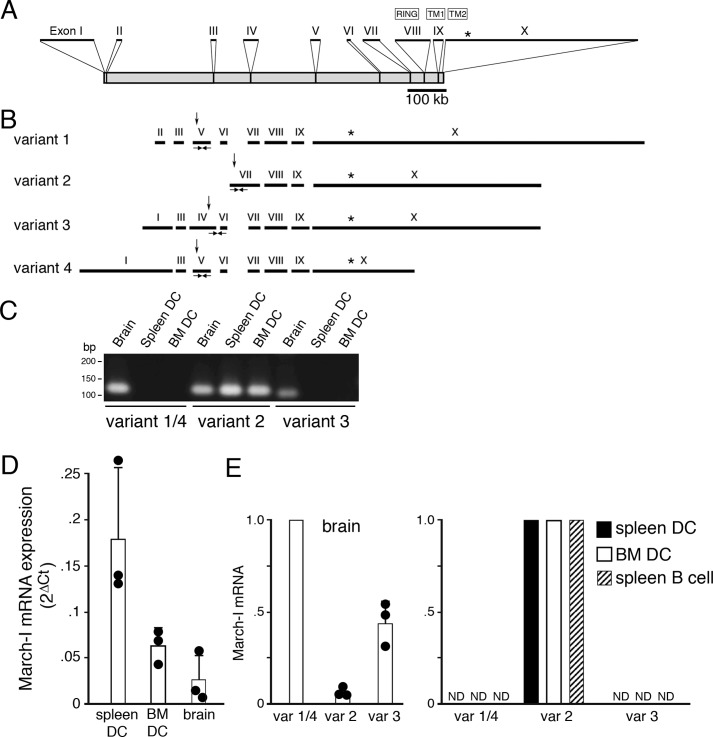
March-I v2 is the predominant March-I variant expressed in DCs. A, schematic representation of the March-I gene as described in UCSC Genome Browser. The positions of each exon (I–X), their relative size, and their location within the March-I gene is indicated. The location of the translation stop codon in exon X is indicated by an asterisk. B, the compositions of March-I variant 1, variant 2, variant 3, and variant 4 (as described in Ensembl) are indicated. The position of the E3 ligase RING domain, transmembrane domain 1 (TM1), and transmembrane domain 2 (TM2) are indicated. The relative position of the translation initiation codon in each variant is indicated by an arrow, and the common translation stop codon in exon X is indicated by an asterisk. The locations of specific PCR primers that specifically amplify March-I fragments specific to variant 1/4, variant 2, and variant 3 are indicated. C–E, PCR primers specific for March-I variant 1/4, variant 2, or variant 3 were used to amplify mRNA from spleen DCs, BM DCs, and brain. C, representative gel showing the presence of March-I variant 2 in all tissues and only small amounts of variant 1/4 and variant 3 in the brain. Forty cycles of PCR amplification for each primer pair were performed, and aliquots of the PCR were analyzed on an agarose gel. D, quantitation of the amount of total March-I mRNA (using a primer set common to all March-I variants) was performed by RT-PCR, and data were normalized to expression of GAPDH in each sample. Data are shown as the 2ΔCt (ΔCt = CtGAPDH − CtMarch-I) value for each condition. The results shown are the average (error bars represent S.D.) of three independent experiments. E, the relative amount of each March-I isoform present in spleen DCs, BM DCs, spleen B cells, and brain was determined by RT-PCR, and the 2ΔCt (ΔCt = CtGAPDH − CtMarch-I) value for each sample was determined. The amount of mRNA for the most abundant March-I variant present in spleen DCs, BM DCs, B cells, and brain was arbitrarily designated a value of 1. The results shown are the average (error bars represent S.D.) of three independent experiments. var, variant; ND, not detectable.