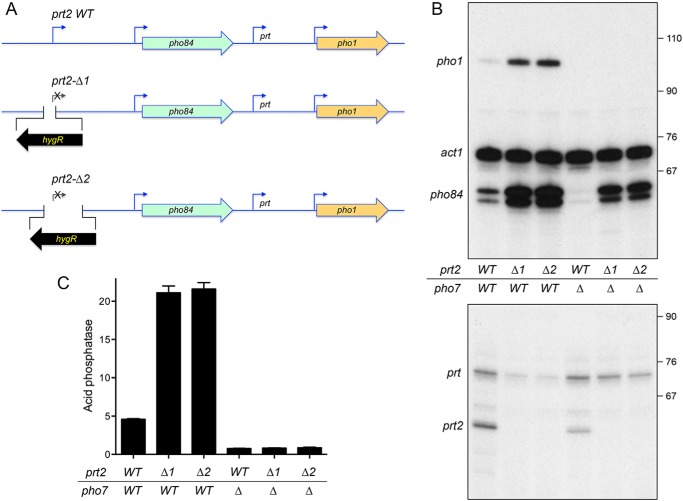
Figure 2.
How insertional inactivation of prt2 affects the expression of downstream genes. A, schematic depiction of the wildtype prt2-pho84-prt-pho1 gene cluster on chromosome II and two prt2-inactivating mutants (Δ1 and Δ2) in which the bracketed segments flanking the prt2 transcription start site were deleted and replaced with an oppositely oriented gene that confers hygromycin resistance (hygR). B, total RNA from fission yeast cells with the indicated prt2 and pho7 genotypes was analyzed by reverse transcription primer extension using a mixture of radiolabeled primers complementary to the pho84, act1, and pho1 mRNAs (top panel) or the prt and prt2 lncRNAs (bottom panel). The reaction products were resolved by denaturing PAGE and visualized by autoradiography. The positions and sizes (nt) of DNA markers are indicated on the right. C, acid phosphatase activity of the indicated strains grown in rich medium was assayed by conversion of p-nitrophenyl phosphate to p-nitrophenol. The y axis specifies the phosphatase activity (A410) normalized to input cells (A600). The error bars denote S.E.