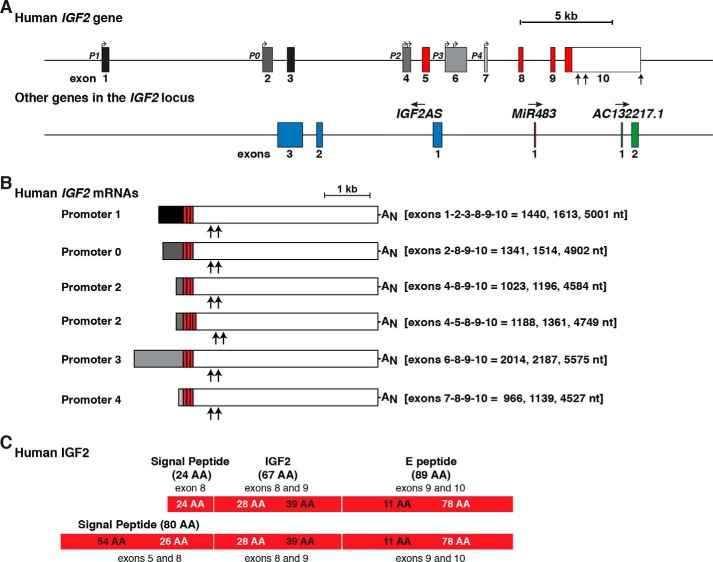
Figure 2.
Human IGF2 gene structure and expression. A, close-up view of the human IGF2 gene, illustrating promoters P0 and P1–P4, and exons 1–10. All 5′-noncoding exons are in black or shades of gray; coding regions are in red, and the 3′ UTR of exon 10 is in white. Bent arrows signify transcription start sites, and vertical arrows delineate different polyadenylation sites in exon 10. Below IGF2 are three other non-coding genes that are embedded within the boundaries of human IGF2. Their direction of transcription is indicated. For all genes, exons are represented as boxes, and introns as horizontal lines. A scale bar is shown. All IGF2AS exons are blue; the MiR483 exon is dark red, and AC132217.1 exons are green. B, diagram of the six major classes of human IGF2 mRNAs. The responsible promoters are listed, with their 5′ non-coding exons in black or gray; coding exons are in red, and the non-coding part of exon 10 is in white to match A. The lengths of each class of mRNAs are indicated in nucleotides (nt), and exons found in each transcript are listed. The locations of polyadenylation sites of shorter transcripts are depicted by vertical arrows (23, 24), with AN representing the polyadenylic acid tail at the 3′ end of the longest mRNAs. C, diagram of the two human IGF2 protein precursors, showing the derivation of each segment from different IGF2 exons. The contributions of different exons to amino acids in different parts of each protein are shown in alternating black and white text. Mature, 67-amino acid IGF2 (center), presumptive signal peptides (left), and the 89-residue COOH-terminal E-peptide (right) are each labeled.