Abstract
Purpose
To identify the genetic cause and describe the phenotype in four families with autosomal recessive retinitis pigmentosa (arRP) that can be associated with pseudocoloboma.
Design
Case series.
Subjects
Seven patients from four unrelated families with arRP of which three patients had bilateral early-onset macular pseudocoloboma.
Methods
We performed homozygosity mapping and whole-exome sequencing (WES) in five probands and two unaffected family members of four unrelated families. Subsequently, Sanger sequencing and segregation analysis were done in additional family members. We reviewed the medical history of individuals carrying IDH3A variants and performed additional ophthalmic examinations, including full-field electroretinography (ffERG), fundus photography, fundus autofluorescence imaging and optical coherence tomography.
Main Outcome Measures
IDH3A variants, age at diagnosis, visual acuity, fundus appearance, visual field, ffERG, fundus autofluorescence and OCT findings.
Results
We identified seven different variants in IDH3A in four unrelated families, i.e. five missense, one nonsense and one frameshift variant. All subjects developed symptoms early in life ranging from night blindness to decreased visual acuity and were diagnosed between the ages of one and 11 years. Four subjects with biallelic IDH3A variants displayed a typical arRP phenotype and three subjects were diagnosed with arRP and pseudocoloboma of the macula.
Conclusions
IDH3A variants were identified as a novel cause of typical arRP, in some individuals associated with macular pseudocoloboma. We observed both phenotypes in two siblings carrying the same compound heterozygous variants, which could be explained by variable disease expression and warrants caution when making assertions about genotype-phenotype correlations.
Introduction
Inherited retinal diseases (IRDs) are a major cause of incurable blindness in children and young adults and are characterized by progressive degeneration of photoreceptor and/or retinal pigment epithelium (RPE) cells. Retinitis pigmentosa (RP) is the most common IRD, with a worldwide prevalence of approximately 1 per 4,000 individuals1–4. In typical RP, patients suffer from night blindness, visual field constriction, and eventually decreased visual acuity5. Characteristic fundus features of RP are bone spicule-like pigmentations, narrowed blood vessels, a waxy, pale appearance of the optic disk, and a relatively preserved macular region.
Leber congenital amaurosis (LCA) is considered the most severe and earliest occurring form of IRD, characterized by severe visual loss, roving eye movements or nystagmus and non-recordable or severely reduced cone and rod electroretinography (ERG) amplitudes within the first year of life6. In practice, the demarcation between LCA and early-onset RP (also called juvenile RP or early onset retinal dystrophy (EORD)) is not always clear and nowadays both disorders are considered as part of the same spectrum due to clinical and molecular overlap.
RP can be inherited in all Mendelian inheritance patterns, and non-Mendelian inheritance patterns such as digenic, mitochondrial or the occurrence of de novo mutations. Eighty-two genes have been associated with this disorder (Retnet; https://sph.uth.edu/retnet/). LCA mainly transmits in an autosomal recessive manner and 22 genes have been identified up to now, of which 12 overlap with early forms of RP. Mutations in these IRD-associated genes reveal various underlying biochemical pathways such as phototransduction, visual cycle regulation, ion homeostasis, cell structure maintenance, ciliary processes, pre-mRNA processing, gene transcription and lipid metabolism. In approximately 60% of LCA and RP cases disease-causing variants can be identified by whole-exome sequencing (WES)7–11. The remaining cases are interesting candidates for the identification of novel disease-associated genes. By sharing WES data and homozygosity mapping data within the European Retinal Disease Consortium (ERDC; 15 research groups in 11 countries), we have been able to identify several new genes underlying IRDs12–15. In this study, we report IDH3A as a novel gene mutated in seven cases with autosomal recessive RP, with early macular involvement and pseudocoloboma.
Methods
Subjects and Clinical Evaluation
The study protocol adhered to the tenets of the Declaration of Helsinki and received approval from the respective local ethics committees. Written informed consent was obtained from all participants or parents of children prior to their inclusion in this study. Patients were ascertained in the Rotterdam Eye Hospital (Rotterdam, The Netherlands), Hadassah-Hebrew University Medical Center (Jerusalem, Israel), Leiden University Medical Center (The Netherlands) or recruited from the IRD registry in the Division of Human Genetics, University of Cape Town (South Africa).
A full medical and family history was taken and retrospective data were retrieved from referring ophthalmologists. Patients underwent a clinical examination including visual acuity testing and retinal imaging. Retinal fundus imaging was obtained by color fundus photography (Topcon Great Britain Ltd, Berkshire, UK). Fundus autofluorescence (FAF) imaging of the posterior pole with fundus-camera-based autofluorescence photography (Topcon Great Britain Ltd, Berkshire, UK) or confocal scanning laser ophthalmoscopy (cSLO) (Spectralis; Heidelberg Engineering Ltd, Heidelberg, Germany) and Spectral domain optical coherence tomography (SD-OCT). Wide angle color and autofluorescence fundus photos were obtained in two patients using an OPTOS system (model 200Tx, OPTOS Plc, Dunfermline, Scotland, United Kingdom). Color vision was evaluated using the Ishihara 38-plate and the Farnsworth D-15 panel. Visual field (VF) testing was performed using a Goldmann kinetic perimeter.
Full-field electroretinography (ffERG) was available from five patients, which incorporated the International Society for Clinical Electrophysiology of Vision (ISCEV) standards. For two Dutch patients from family A DTL-electrodes (Diagnosys, Cambridge, UK) (patient A-II:1) and skin electrodes (patient A-II:2) were used in combination with a computerized system (Espion Ganzfeld, Diagnosys, Cambridge, UK). Light and dark-adapted responses were measured in accordance with ISCEV extended protocol. The dark-adapted rod and mixed responses were acquired after 20 minutes of dark adaptation; responses to different intensities were recorded from 0.0001 to 22 cds/m2 in 12 incremental steps. Under light adapted conditions the 30-Hz flicker and single flash responses from 0.3 to 10 cds/m2 in four incremental steps were recorded under a background light of 30cd/m2. Additionally, we performed S-cone specific testing by using an amber stimulus on a blue background and a blue stimulus on an amber background after one minute of dark adaptation. All ERG responses were acquired at a sample rate of 2000Hz and multiple sweeps were averaged. For two Israeli patients from family B ffERG was performed using monopolar corneal electrodes (Henkes type; Medical Workshop B.V., Groningen, The Netherlands) and a computerized system (UTAS 3000; LKC, Gaithersburg, MD, USA). Cone responses to 30-Hz flashes of white light were acquired under a background light of 21 cd/m2. Dark-adapted responses, including a rod response to a dim blue flash and a mixed cone-rod response to a standard white flash, were acquired after 30 to 45 minutes of dark adaptation. Between two and four sets of responses were recorded in each condition to verify repeatability. All ERG responses were filtered at 0.3 to 500 Hz, and signal averaging was applied. Responses were evaluated using local reference values.
Genetic analysis
Genomic DNA was isolated from peripheral blood lymphocytes according to standard procedures. WES of members of families A, B and C was performed using Agilent’s SureSelect All Human Exome version 4 kit (Agilent Technologies, Santa Clara, CA, USA), followed by sequencing on a SOLiD4 sequencing platform (Life Technologies, Carlsbad, CA, USA) with an average depth of 80X. Exome capture for family D was performed using the Nextera Rapid Capture Expanded Exome kit (Illumina, San Diego, CA, USA), and sequenced on a HiSeq2500 platform (Illumina). Reads were subsequently mapped against the UCSC genome browser’s Human hg19 assembly (build 37) and variants were called using the Genome analysis Toolkit (GATK) v2 according to the recommended guidelines (https://software.broadinstitute.org/gatk/best-practices/).
Homozygosity mapping
Runs of homozygosity were determined using the online tool Homozygosity Mapper, using variant call format (*.vcf) file as an input file. Homozygous regions larger than 4 Mb were selected and ranked. Assuming autosomal recessive inheritance, we used the identity by descent (IBD) regions to narrow down candidate disease-associated variants identified from the WES data.
Sanger sequencing
Primers for amplification of coding exons and flanking exon-intron boundaries of IDH3A (NM_005530.2) were designed with Primer3 plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). Primer sequences and PCR conditions are available upon request.
Variant prioritization
Variants identified in the WES data of probands A-II:2, B-II:1, C-II:1 and D-II:1 were filtered as follows: (1) Selection of variants with a minor allele frequency (MAF) ≤0.5% in a stringent and ≤1.5% in a less stringent filtering. MAFs were obtained from the Exome Aggregation Consortium (ExAC), dbSNP and in-house variant databases; (2) Classification of variants based on their effect (nonsense, frameshift, missense or splice site); (3) Selection of variants that occurred in homozygous or compound heterozygous state; (4) Assess the pathogenicity of candidate variants using the Combined Annotation Dependent Depletion (CADD) (http://cadd.gs.washington.edu/) score. This program incorporates several widely used in silico tools, such as SIFT (http://sift.jcvi.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), Mutation Taster (http://www.mutationtaster.org/), PhyloP and Grantham score. SIFT predicts whether an amino acid substitution affects protein function, PolyPhen-2 predicts whether an amino acid substitution affects protein structure and function, and Mutation taster estimates the impact of a variant on the protein and the gene product. PhyloP scores measure evolutionary nucleotide conservation at individual alignment sites; positive scores are associated with conserved regions compared to the evolution expected under neutral drift. Finally, the Grantham score represents the evolutionary distance between two amino acids, the more distant two amino acids are, the more damaging their substitution is.
Results
Clinical findings
We ascertained seven patients from four families, between the ages of 3 to 26 years. Their clinical characteristics are summarized in Table 1 and the family pedigrees are shown in Figure 1. All patients had early onset night blindness and were diagnosed between the ages of 1 and 11 years (y). Visual acuity (VA) was available for all patients and ranged from 20/20 to counting fingers (CF) at 1 meter.
Table 1.
Clinical characteristics of patients with biallelic IDH3A variants.
| ID gender age last visit |
Origin | Variants | Age of onset | Refractive error, spherical equivalent (age) | Initial symptoms | Snellen visual acuity (age) | Fundus | Electroretinography (age) | Perimetry (age) | OCT (age) | Fundus autofluoresence (age) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A-II:1 female 7 y |
the Netherlands | c.403del p.(Thr135Profs*6) |
5 | RE −0.50D LE −0.50D (7) |
Nyctalopia | RE 10/20 LE 8/20 BE 10/20 (7) |
Bilateral pink optic discs and mild attenuated blood vessels. Broad foveal reflexes. RPE alterations in the fovea and periphery. | DA: no isolated rod responses,
small mixed responses with normal latencies LA: 30Hz no responses, small single flash response with delayed b-wave (6) $ |
Peripheral constriction and sensitivity loss with V4 target (6) | Bilateral disruption of the ellipsoid zone in the foveola (6) | Intact autofluorescence of the posterior pole with fundus-camera based autofluorescence (6) & |
| c.716T>C p.(Met239Thr) | |||||||||||
|
| |||||||||||
| A-II:2 male 3 y |
the Netherlands | c.403del p.(Thr135Profs*6) |
1 | RE +0.50D LE +1.00D (2) |
Strabismus, nyctalopia, impaired visual acuity | RE 2/20 LE 3/20 BE 8/20 (3) |
Bilateral pink optic discs, attenuated vessels, distinct atrophy of the RPE and choriocapillaris demarcated by a hyperpigmented ring in the macular region, peripheral RPE alterations. | NR (3) * | NP | Atrophy of the RPE, abnormal outer retinal layers (3) | NP |
| c.716T>C p.(Met239Thr) |
RE 3/20 LE 3/20 BE 3/20 (5) |
||||||||||
|
| |||||||||||
| B-II:1 male 18 y |
Israel | c.911C>A p.(Pro304His) |
11 | BE +3.50D (11) | Nyctalopia | BE 20/20 (11–16) | Hypermetropic optic discs, normal retinal blood vessels, CME, and BSP associated with grayish RPE atrophy in the midperiphery of the retina LE>RE. | DA: NR LA: markedly reduced and delayed 30Hz cone flicker responses (12) |
Goldmann VFs mildly constricted superiorly and nasally LE>RE with IV4e target (18) | Severe cystoid macular edema (RE>LE) with thickening of the retina and loss of normal foveal structure, thinning and loss of the photoreceptor ONL in the para-macular regions (18) | Enlarged central hypoautofluorescent area surrounded by a faint hyperautofluorescent ring (18) |
| c.938T>C p.(Met313Thr) |
RE +2.25D LE +2.00D (15) |
||||||||||
|
| |||||||||||
| B-II:2 female 15 y |
Israel | c.911C>A p.(Pro304His) |
11 | BE −3.75D (13) | Nyctalopia | BE 20/20 (11–16) | Normal-appearing optic discs and blood vessels, an abnormal macular reflex without CME, and grayish discoloration of the RPE in the midperiphery associated with salt and pepper changes as well as mild BSP changes nasal>temporal. | DA: no isolated rod responses,
significant mixed rod-cone responses to a standard white
flash LA: moderately reduced and delayed 30Hz cone flicker responses (12) |
Goldmann VFs preserved for IV4e target in both eyes (15) | Presence of the photoreceptor nuclei layer in the foveas without edema, but marked thinning and loss of the ONL is evident in para-macular regions (15) | Prominent hyperautofluorescent ring surrounds the center of the maculas in BE (15) |
| c.938T>C p.(Met313Thr) | |||||||||||
|
| |||||||||||
| C-II:1 female 20 y |
the Netherlands | c.524C>T p.(Ala175Val) |
10 | BE −2.00D (20) | Nyctalopia | BE 16/20 LE 18/20 (20) |
Pale (grayish) aspect of the optic disc, attenuated blood vessels, broad foveal reflex with orange coloured aureola around the foveola, RPE alterations from midperiphery towards far periphery. | DA: almost NR LA: 30 Hz cone flicker responses recognizable, single flash responses significantly reduced and delayed (14) |
Goldmann VFs severe peripheral constriction (<10 degrees) with sensitivity loss (20) | Extensive peripheral degeneration, distortion of the macular architecture (20) | NP |
| c.524C>T p.(Ala175Val) | |||||||||||
|
| |||||||||||
| D-II:2 female 26 y (deceased) |
South Africa | c.463G>T p.(Gly155*) |
8 | NP | Nyctalopia, impaired visual acuity | BE CF 1m (26) | Optic atrophy, mild arteriolar narrowing, severe macular pigmentation, pseudocoloboma of the macula and severe mid peripheral bone spicule pigmentary changes. | NP | NP | NP | NP |
| c.946C>T p.(Arg316Cys) | |||||||||||
|
| |||||||||||
| D-II:3 female 26 y |
South Africa | c.463G>T p.(Gly155*) |
8 | NP | Nyctalopia, impaired visual acuity | BE CF 1m (23) | Optic atrophy, mild arteriolar narrowing, severe macular pigmentation, coloboma-like atrophy of the macula and severe mid peripheral bone spicule pigmentary changes. | NP | NP | Macular atrophy and macular pseudocoloboma, showing lamellar disorganization (lamellar structures were visible but could not be identified) (26) | Strongly reduced autofluorescence within the macula, diffusely increasing in a broad ring around the macula, and fading towards the periphery (26) |
| c.946C>T p.(Arg316Cys) | |||||||||||
DTL electrodes,
skin electrodes,
fundus-camera-based autofluorescence photography Topcon TRC-50EX/50IX (Megaplus2), BE: both eyes, BSP: bone spicule-like pigmentation, CF: counting fingers, CME: cystoid macular edema, D: diopters, DA: dark adapted, LA: light adapted, LE: left eye, NR: no response, NP: not performed, ONL: outer nuclear layer, RE: right eye, RPE: retinal pigment epithelium, VF: visual field.
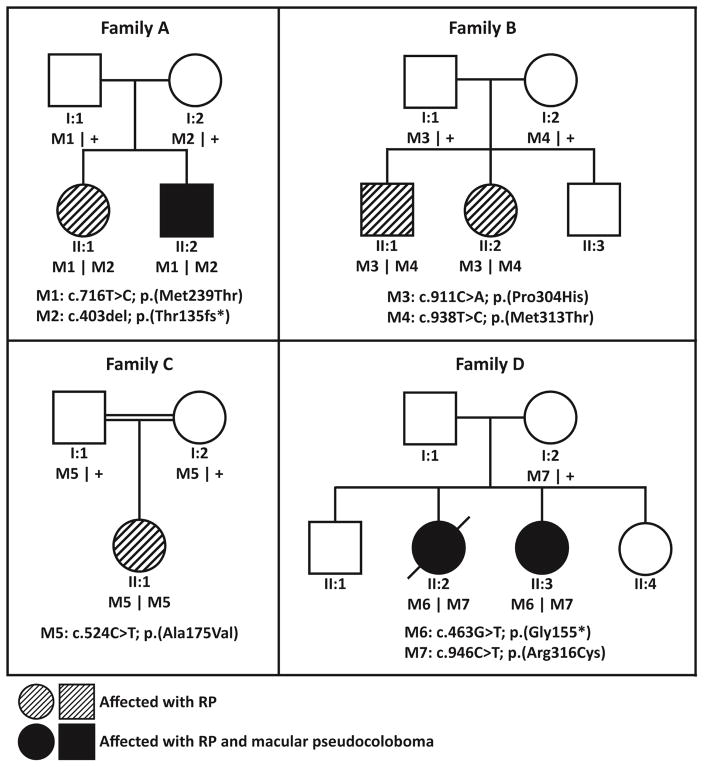
Figure 1.
Pedigrees of families included in this study.
The youngest patient (A-II:2) of a Dutch family was diagnosed with a macular pseudocoloboma at the age of 1 y and was referred to the Rotterdam Eye Hospital with decreased VA and strabismus at 3 y of age. At that stage, VA was CF with a fine horizontal nystagmus. On fundus examination, a macular pseudocoloboma was noted with peripheral RPE alterations (Figure 2D). The dimensions of the pseudocoloboma appeared unchanged and VA remained stable at age 5 y. The OCT scan showed severe atrophy of the outer retinal layers and the RPE at the colobomatous lesion (Figure 2E). With ERG testing no responses could be distinguished from noise level under light-and dark-adapted circumstances. His family history revealed that his sister (A-II:1), who was 3 years older, suffered from night blindness. Although her visual acuity was still normal, RPE alterations were noted at her macula, and OCT scans showed subfoveal alterations of the ellipsoid zone, confirming early macular involvement (Figures 2A, B). The fundus camera-based autofluorescence image appears dark because of underexposure with a very low sensitivity setting during acquisition; the autofluorescence of the posterior pole looks intact. Fundus examination also revealed early signs of peripheral degeneration, and ERG and Goldman visual field testing were compatible with the diagnosis of RP.
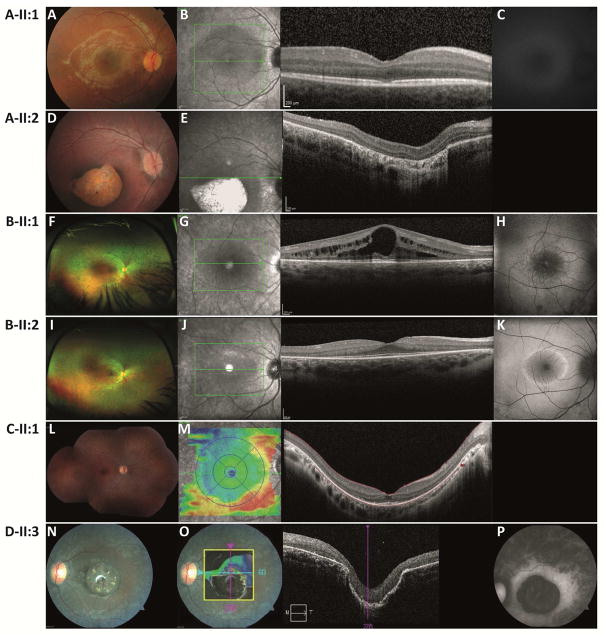
Figure 2.
Retinal imaging data for patients with IDH3A variants.
(Wide-field) color fundus photographs, optical coherence tomography OCT and fundus autofluorescence (FAF) imaging with fundus-camera-based autofluorescence photography or confocal scanning laser ophthalmoscopy (cSLO).
Patient A-II:1 at age 7 y, (A) pink optic discs, mildly attenuated blood vessels, RPE alterations in the fovea and periphery, (B) small central disruptions in the ellipsoid zone, (C) intact autofluorescence of the posterior pole.
Patient A-II:2 at age 3 y, (D) distinct atrophy of the RPE and choriocapillaris in the macular region demarcated by a hyperpigmented ring, peripheral RPE alterations, (E) atrophy of the RPE, abnormal outer retinal layers.
Patient B-II:1 at age 18 y, (F) mid-peripheral atrophy with bone spicule-like pigmentation nasal>temporal, OS>OD, (G) bilateral severe cystoid macular edema (RE>LE) with thickening of the retina and loss of normal foveal structure, thinning and loss of the photoreceptor nuclei layer (ONL) is evident in the para-macular regions of the scans, (H) hyper-autofluorescent ring around the fovea.
Patient B-II:2 at age 15 y, (I) mid-peripheral atrophy with bone spicule-like pigmentation nasal>temporal, OS>OD, (J) presence of the photoreceptor nuclei layer in the fovea without edema, but marked thinning and loss of the ONL is evident in para-macular regions, (K) prominent hyper-autofluorescent ring surrounding the center of the macula.
Patient C-II:1 at age 20 y, (L) pale aspect of the optic disc, attenuated blood vessels, RPE alterations from mid-periphery towards the far periphery, (M) extensive peripheral atrophy of the outer segments and distortion of the foveal architecture.
Patient D-II:2 at age 26 y, (N) peripheral bone-spicule pigmentary changes, peripheral and mid-peripheral pigmentary retinopathy, macular pigmentation with severe pseudocoloboma-like atrophy, mild arteriolar narrowing and mild optic atrophy, (O) macular atrophy with lamellar disorganization, (P) strongly reduced autofluorescence within the macula, diffusely increasing in a broad ring around the macula, and fading towards the periphery.
The Israeli family included two affected siblings, an older brother and a younger sister (Figure 1, Table 2), who were evaluated at age 11 y, based on suspicion of night blindness. Best-corrected visual acuity (BCVA) in the eldest sibling (B-II:1) was well preserved up until his last exam at 18 y of age (20/20 in both eyes with hyperopia), despite severe bilateral cystoid macular edema (CME) (Figures 2G, H) that was noted already at age 12 y. CME was treated although visual acuity was optimal but remained refractory to multiple trials of treatment with systemic and topical carbonic anhydrase inhibitors as well as topical Voltaren® drops. On fundus examination mid -peripheral bone spicule-like pigmentations and atrophy were noted (Figure 2F), ffERG testing and Goldman visual field testing were compatible with his symptoms and the RP phenotype. Color vision testing using the Ishihara 38-plate and the Farnsworth D-15 panel was within normal limits. OCT and fundus autofluorescence imaging reflect CME and peripheral atrophy of the outer segments (Figures 2G, H). BCVA in his younger sister (B-II:2) was also preserved up until her last exam at age 15 y (20/20 in both eyes), with myopic correction. Fundus findings (Figure 2I) include normal-appearing optic discs and blood vessels, an abnormal macular reflex without CME, and subtle RPE alterations with mild bone spicule-like pigmentations nasal>temporal in the mid-periphery. ffERG testing revealed better responses than those observed in her brother at the same age (Table 1). Goldmann visual fields to a IV4e target were preserved and color vision was normal. OCT imaging showed that the outer nuclear layer was still present in the foveal area, but markedly thins out in parafoveal regions (Figure 2J). An hyper-autofluorescent ring was seen in the maculas and hypo-autofluorescent patches were present beyond the vascular arcades (Figure 2F).
Table 2.
IDH3A variants identified in this study and their in silico functional analyses.
| DNA variant | Protein variant | dbSNP id | ExAC allele frequency (%) | SIFT | Polyphen-2 | PhyloP | Grantham Score | CADD Phred |
|---|---|---|---|---|---|---|---|---|
| c.403del | p.(Thr135fs*) | N.A. | 0 | N.A. | N.A. | N.A. | N.A. | N.A. |
| c.463G>T | p.(Gly155*) | N.A. | 0 | N.A. | N.A. | N.A. | N.A. | 38 |
| c.524C>T | p.(Ala175Val) | rs765473830 | 0.0008 | Deleterious | Probably damaging | 5.9 | 64 | 34 |
| c.716T>C | p.(Met239Thr) | N.A. | 0 | Deleterious | Possibly damaging | 4.97 | 81 | 14.32 |
| c.911C>A | p.(Pro304His) | rs756712426 | 0.0008 | Deleterious | Probably damaging | 5.94 | 77 | 33 |
| c.938T>C | p.(Met313Thr) | rs149862950 | 0.0002 | Deleterious | Probably damaging | 4.97 | 81 | 23.5 |
| c.946C>T | p.(Arg316Cys) | rs770798851 | 0.0008 | Deleterious | Probably damaging | 4.32 | 180 | 28.9 |
An isolated case of Dutch descent (C-II:1) was referred to the Leiden University Medical Center with complaints of night blindness at the age of 10 y. BCVA was suboptimal at last examination (OD 16/20, OS 18/20) with myopic correction. On fundus examination, typical RP features were present (Figure 2L). ffERG showed severely reduced dark- and light-adapted responses. Visual fields were severely constricted (10° radius). On OCT imaging extensive peripheral degeneration was seen with distortion of the macular alterations of the outer segments and RPE architecture (Figure 2M).
Two South African sisters (D-II:2, D-II:3) (Figure 1) had an onset age of 8 y with complaints of night blindness and decreased visual acuity. In their early twenties, both were diagnosed with a rapid progressive form RP and macular pseudocoloboma. Their vision at that stage was limited to counting fingers at 1 meter. Fundus examination revealed severe mid-peripheral bone spicule pigmentary changes, mild arteriolar narrowing, pseudocoloboma of the macula and optic atrophy (Figure 2N). The youngest sister was examined 4 years later (Figure 2N). At that time the oldest sister had died due to an unrelated infection. The fundus autofluorescence was strongly reduced within the macula (Figure 2P), the optic disk appears white as result of an inaccurate sensitivity setting during acquisition. The OCT showed macular atrophy and confirmed the pseudocoloboma, also showing lamellar disorganization (Figure 2O).
The disorder did not seem to be associated with distinct hypermetropia or myopia, early-onset cataract, or keratoconus. Both the deceased patient from South Africa and the male patient from Israel developed edema of the lower extremities. In the South African patient the bilateral edema was thought to be caused by low serum albumin, but in the Israeli patient no cause could be appointed for the unilateral edema after a thorough work-up. The significance of this is unknown and no additional systemic disorders were detected, nor any systemic associations.
Genetic findings
WES revealed biallelic variants in IDH3A in seven patients from four families, and are summarized in Tables 1 and 2. All variants were novel and no other pathogenic variants were found in other genes after WES in families A and B.
Sequencing of nicotinamide nucleotide adenylyltransferase 1 (NMNAT1), a candidate gene for this phenotype, in subject A-II:2, did not reveal a suspected disease-causing variant and neither did targeted sequencing of a panel of 256 genes associated with inherited vision disorders, including known IRD-associated genes. Subsequently, analysis of the complete exome was performed and two variants in IDH3A were found. No alternative candidate variants remained after filtering. Additional genetic testing of his sister and parents showed that the variants segregated with the disease phenotype (Figure 1). Screening of IDH3A in WES data from 121 isolated unsolved RP/arRP cases did not reveal additional cases with IDH3A variants and IBD data from 199 unsolved isolated RP/arRP cases did not reveal homozygous regions > 4 Mb that spanned the IDH3A gene.
In parallel, a genetic analysis of Family B (MOL0655), which included Sanger sequencing of founder mutations previously identified in RP patients of Jewish origin, followed by trio WES analysis (two affected siblings and their mother), revealed compound heterozygous variants in IDH3A (p.(Pro304His); p.(Met313Thr)) in subjects B-II:1 and B-II:2. No alternative candidate variants remained after filtering. Screening the IDH3A variants in a set of 261 ethnically-matched Israeli index cases with RP did not reveal other cases with these variants. In addition, we scrutinized homozygosity mapping data of 193 families. Twelve showed homozygosity of the IDH3A region but Sanger sequencing of the exons of IDH3A revealed no additional variants. This family was part of a cohort of 138 families with isolated RP/arRP in which WES was performed and led to a molecular diagnosis was in 56 families.
Using Homozygosity Mapper, we identified eight homozygous regions larger than 30 Mb in patient C-II:2. WES data analysis did not yield any variants in IRD genes that are located within this region. Upon stringent filtering step, two homozygous missense changes, i.e. in IDH3A (p.(Ala175Val) and TSPAN3 (p.(Arg80Cys)), remained as candidate variants (Table 2). The TSPAN3 variant, p.(Arg80Cys), had a minor allele frequency of 0.0017 in the ExAC database and was predicted to be benign by SIFT and PolyPhen2 in silico prediction tools, but had a high PhyloP (3.4), Grantham (180) and CADD-Phred score (15.70). Northern blot analysis and data from an expressed sequence tag (EST) database demonstrated a ubiquitous gene expression16.
Subject D-II:1 was previously analyzed using an ABCA4 Microarray from Asper Biotech (558 variants) using arrayed-primer extension technology,17 and subsequently using an LCA Microarray 17 (780 variants in 15 LCA-associated genes), both revealing no causal variants. Exome sequencing of a panel of 217 known IRD-associated genes was negative18, but analysis of ERDC candidate genes (http://www.erdc.info/candidateirdgenes) revealed biallelic variants in IDH3A which segregated with the disease phenotype (Figure 1). Screening of these variants in a set of 16 indigenous African families (n=56 individuals) with IRDs was negative.
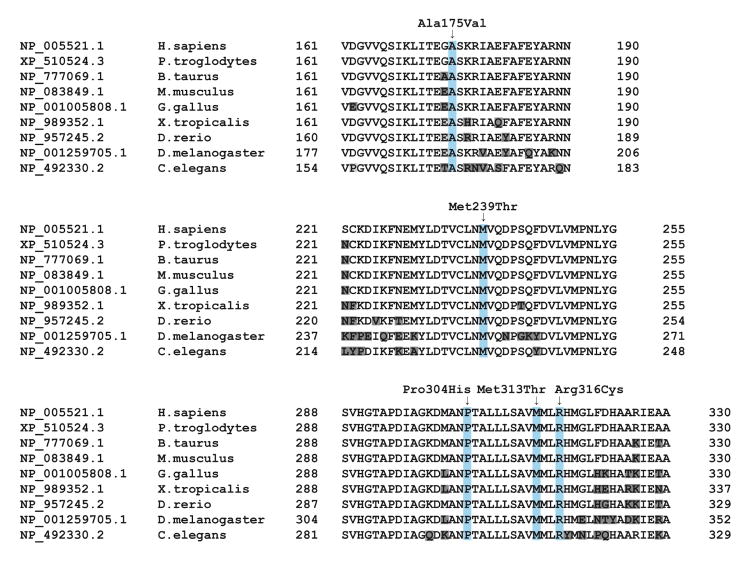
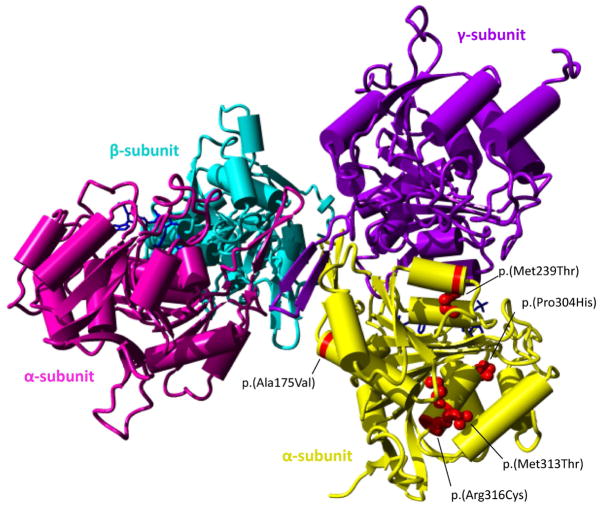
In total, seven different variants were detected in IDH3A in our cases. The resulting transcripts of the two nonsense variants (p.(Thr135fs*), p.(Gly155*)) are predicted to undergo nonsense mediated decay, suggesting that these variants are true loss-of-function alleles. All missense variants (p.(Ala175Val), p.(Met239Thr), p.(Pro304His), p.(Met313Thr), p.(Arg316Cys)) were indicated as pathogenic by in silico prediction tools (Table 2). All missense variants were located at conserved residues of the IDH3A protein (Figure 3), and fall within the conserved mitochondrial NAD-dependent IDH domain - a conserved nucleotide binding domain typical of NAD- or NADP-dependent dehydrogenases (NCBI’s conserved domain database). The p.(Ala175Val) variant is located in a region that is in close contact with other IDH3 subunit(s). The impact of the variant on the structure is minor; however due to its critical location, it may disrupt the binding between subunits to form the IDH3 complex as the valine amino acid is slightly bulkier than alanine (Figure 4). The p.(Met239Thr) missense variant affects an amino acid residue buried in the core close to the interaction surface with the IDH3 gamma subunit (Figure 4). The smaller size of threonine will disturb the stabilizing hydrophobic interactions and might affect subunit interaction. The p.(Pro304His) missense variant might affect the interaction with the NADH ligand (Figure 4). The proline is predicted to be located closely to the NADH ligand, which allows them to interact. The rigidity of proline provides a stable structure for the interaction with the ligand. Hence, this alteration may destabilize and deform the rigid structure, and affect the interaction with the ligand. Moreover, histidine is a large amino acid, which may also alter the van der Waals atomic contacts with other amino acids nearby. The p.(Met313Thr) missense variant has no predicted impact on the protein structure but changes the hydrophobic amino acid to polar. Thus any hydrophobic activity needed in this position will be reduced. Lastly, the p.(Arg316Cys) missense variant is located in the same helix as mutant p.(Met313Thr). However, the arginine sidechain is located on the surface where it can make stabilizing ionic interactions with glutamic acid 328. The insertion of cysteine at this position does not have a big effect on the size of the amino acid at this position, but the stabilizing interaction in this domain will be affected. Additionally, a reactive sulfur atom will appear on the surface of the protein.
Figure 3.
Multiple sequence alignment of IDH3A orthologs around missense variant sites (highlighted in light blue) generated by NCBI HomoloGene. Amino acids highlighted in grey are not identical to the corresponding amino acid in the human ortholog.
Figure 4.
3D structure of the IDH3 complex and the location of the missense variants.
The IDH3 protein complex contains two alpha subunits (pink, yellow), one beta subunit (purple) and one gamma subunit (blue). Missense variants are indicated in red in only one of the two alpha subunits for clarity. In patients with compound heterozygous variants, the NAD-IDH complex may consist of two identical alpha subunits altered by the same variant or two different alpha subunits.
Discussion
In this study, we identified novel variants in IDH3A in four cases with typical arRP and three cases with arRP and macular pseudocoloboma from four unrelated families. These families originated from the Netherlands, Israel, and South Africa. All patients had visual complaints early in life ranging from night blindness to severe visual impairment. In patients with RP and macular pseudocoloboma (A-II:2, D-II:2 and D-II:3) the visual impairment was more severe than in patients with isolated RP (A-II:1, B-II:1, B-II:2 and C-II:1), which might explain why they were diagnosed at a younger age. The youngest patient with RP and pseudocoloboma (A-II:2) was three, whereas his sister (A-II:1) of six displayed only some RPE alterations in the macular region with abnormalities in the ellipsoid zone on OCT examination with moderate reduced visual acuity (Table 1, Figure 2B). In two Israeli siblings (B-II:1 and B-II:2) without pseudocoloboma, the visual acuity and visual field remained optimal up until the last examination at age 16, despite bilateral severe CME in B-II:1 (Figure 2G). In both siblings, the ellipsoid zone appeared normal in the macular region on OCT examination (Figure 2J).
IDH3A encodes the mitochondrial NAD+-specific IDHα subunit (IDH3A), a 339 amino acid long protein that is a part of the heterotetramer IDH3, comprised of two α subunits, one β subunit (the product of the IDH3B gene) and one γ subunit (the product of the IDH3G gene)19. IDH3 catalyzes the oxidation of isocitrate to α-ketoglutarate in the citric acid cycle, also known as the Krebs cycle. Recently, a homozygous missense IDH3A variant, p.(Pro304His) was found in a patient suffering from severe infantile encephalopathy with peripheral and autonomic nervous system involvement20. Funduscopy at the age of 8 months revealed RP and bilateral optic atrophy. Functional studies in yeast confirmed the high evolutionary conservation of the IDH3 enzyme. Wild-type human IDH3A complemented the growth defect observed in IDH1- and IDH2-depleted yeast, but human IDH3A harboring the p.(Pro304His) variant could not complement yeast IDH1 and IDH2 function. This suggested that the variant has a significant impact on the function of IDH3A. The same variant is reported here by us to cause nonsyndromic RP in a compound heterozygous state with the p.(Met313Thr) variant in two siblings. The phenotypic difference (severe infantile encephalopathy versus Nonsyndromic RP) might indicate that the p.(Pro304His) variant is responsible for a more severe and profound phenotype, while the p.(Met313Thr) variant is milder and limits the phenotype to the retina. Such correlations were reported in other RP-associated genes including USH2A and some of the genes causing Bardet-Biedl syndrome21–24. Additionally, another group reported on a homozygous Idh3a missense variant, p.(Glu229Lys), in mice with an isolated RP phenotype25. This mutation was found in a large N-ethyl-N-nitrosourea mutagenesis screen.
Biallelic variants in IDH3B, coding for the IDHβ subunit, were previously described by Hartong et al. in three cases from two families with arRP26. A combination of expression analysis and direct sequencing revealed a homozygous frameshift variant that segregated with the disease. Screening of IDH3B in 546 unsolved cases with RP led to the discovery of a second homozygous missense variant. Subsequent functional studies showed a significant decrease in NAD-IDH activity in lymphoblast cell lysates from homozygous patients, compared with wildtype controls and heterozygous carriers. All patients with IDH3B variants presented with RP without macular pseudocoloboma or systemic involvement. As IDH3A codes for the two alpha subunits in the NAD-IDH enzyme, it is plausible that variants affecting the IDH3A protein influence NAD-IDH function in a similar manner as do IDH3B variants. There have been no reports on disease associations for the gene encoding the third component of the NAD-IDH enzyme, IDH3G, but it can be an interesting candidate gene for IRDs.
We deemed the homozygous TSPAN3 variant found in family C with homozygosity mapping less likely to be causal. Currently, there is no retinal disorder or retinal pathway associated with variants in TSPAN3. In addition, the mutant cysteine residue at position 80 is a wild-type amino acid in Tetraodon nigroviridis.
In our IDH3A cohort, patients carried two missense variants or a combination of a missense and a null variant. Some missense variants might be hypomorphic variants that allow for residual IDH3A activity. Remarkably, all patients with arRP and macular pseudocoloboma carried a null variant, which could explain the more severe phenotype. However, one sibling with only mild foveal abnormalities carried the same null and missense variant as her brother who had a macular pseudocoloboma. This difference in phenotype could be caused by differences in IDH3A expression between the siblings. A higher expression of the mutant IDH3A carrying the missense variant in the typical arRP sibling could potentially result in the milder phenotype. But, the NAD-IDH protein complex is subject to strict stoichiometric rules, so it is also possible that different expression levels of IDH3B and IDH3G influence the phenotypic outcome.
RPE abnormalities at the macula at an early stage are more frequently observed in IRDs caused by mutations in genes associated with Leber congenital amaurosis and early-onset RP (e.g. CRB1). Rarely noted is a macular pseudocoloboma, a somewhat uncomfortable term used to describe early and distinct degeneration of a normally appearing macula. ‘Macular coloboma’ is often misused for describing atrophic lesions in the central retina, where true coloboma are the result of failed or incomplete closure of the embryonic fissure during the sixth and seventh week of fetal development and are often associated with microphthalmos or other closure defects. Macular pseudocoloboma was first described by Clausen in 1921 in a family with a dominant inheritance pattern and no systemic involvement27. They were also mentioned in Foxman’s classification of congenital and early-onset RP as a ‘common’ finding in patients with uncomplicated (or non-syndromic) LCA6. Since molecular diagnostics has become more widely available we acknowledge that mutations in different genes involved in various pathways underlying genetic vision disorders can be causal. Pseudocolobomas are characteristic for nicotinamide nucleotide adenylyltransferase 1 NMNAT1-associated LCA28–31, DEAH box polypeptide 38 (DHX38)-associated RP32, and North Carolina type macular dystrophy (NCMD)33, 34, all in which macular atrophy is caused by early-onset neurodegeneration28.
NMNAT1 is an essential enzyme for regenerating the nuclear pool of NAD+ in all nucleated cells in the body and plays a role in neuroprotection28–31. Pseudocolobomas in patients with NMNAT1-associated LCA are the result of retinal degeneration after normal anatomical development, in contrast to true coloboma where the retinal structure was abnormally developed in utero. Mice studies support these findings and show that photoreceptors are extremely susceptible to NMNAT1 dysfunction as their metabolic activity is very high35. The need for NAD+ is critical after light exposure and cannot be met when enzymatic NMNAT1 activity is disrupted28. RPE cells and inner retinal cells undergo degeneration to a lesser extent35.
A homozygous variant in DHX38 was found in a Pakistani patient with early-onset RP and macular pseudocoloboma32. DHX38 encodes the pre-mRNA splicing factor PRP16, that is involved in catalyzing the second step in pre-mRNA splicing. Splicing factor genes are widely expressed, but mutations in some of these genes, such as PRPF3, PRPF8, PRPF31 and SNRNP2000, exclusively result in retinal degeneration without other systemic manifestations.33, 3436, 373838 32, 39
All genes associated with macular pseudocoloboma are widely expressed and are involved in different pathways, such as energy metabolism (IDH3A, NMNAT1), pre-mRNA splicing (DHX38) and transcription control (PRDM13). However, the disease manifestation seems to be restricted to the retina. Possible explanations might be the exceptionally high metabolic turnover in photoreceptor cells, which requires significantly more energy and higher levels of pre-mRNA splicing molecules compared with other cell types. Alternatively, other factors or pathways may compensate for this high consumption in other cell types. There are no reports on IRD patients with two null variants in IDH3A, NMNAT1, and DHX38, which leads us to believe that complete inactivity of these proteins might be incompatible with life.
In conclusion, we identified seven (likely) pathogenic variants in IDH3A leading to typical arRP and arRP associated with macular pseudocoloboma in seven patients from four unrelated families. Thereby we have implicated a second subunit of the IDH3 protein complex to play a crucial role in the human retina.
Acknowledgments
The authors thank patients and family members for participation. LP is supported by Foundation Combined Ophthalmic Research Rotterdam (CORR). The research of GDNA was supported by the Directorate General for Higher Education (DIKTI) of the Ministry for National Education of Indonesia and the Radboud University Medical Center, Nijmegen, The Netherlands. Research in South Africa was funded by Retina South Africa, the South African Medical Research Council and the National Research Foundation of South Africa. Research in Israeli was supported by the United States – Israel Binational Science Foundation (grant number 2011202, to D.S. and A.S.), Foundation Fighting Blindness USA (BR-GE-0214-0639 to D.S. and E.B.), and the Yedidut Research Grant (to E.B.). WES analysis of Israeli and South African samples was supported by Intramural Research program of the National Eye Institute, NIH, USA (EY000546).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Haim M. Epidemiology of retinitis pigmentosa in Denmark. Acta Ophthalmol Scand Suppl. 2002;(233):1–34. doi: 10.1046/j.1395-3907.2002.00001.x. [DOI] [PubMed] [Google Scholar]
- 2.Rosner M, Hefetz L, Abraham FA. The prevalence of retinitis pigmentosa and congenital stationary night blindness in Israel. Am J Ophthalmol. 1993;116(3):373–4. doi: 10.1016/s0002-9394(14)71358-3. [DOI] [PubMed] [Google Scholar]
- 3.Grondahl J. Estimation of prognosis and prevalence of retinitis pigmentosa and Ushersyndrome in Norway. Clin Genet. 1987;31(4):255–64. doi: 10.1111/j.1399-0004.1987.tb02804.x. [DOI] [PubMed] [Google Scholar]
- 4.Bunker CH, Berson EL, Bromley WC, et al. Prevalence of retinitis pigmentosa in Maine. Am J Ophthalmol. 1984;97(3):357–65. doi: 10.1016/0002-9394(84)90636-6. [DOI] [PubMed] [Google Scholar]
- 5.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368(9549):1795–809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 6.Foxman SG, Heckenlively JR, Bateman JB, Wirtschafter JD. Classification of congenital and early onset retinitis pigmentosa. Arch Ophthalmol. 1985;103(10):1502–6. doi: 10.1001/archopht.1985.01050100078023. [DOI] [PubMed] [Google Scholar]
- 7.Beryozkin A, Shevah E, Kimchi A, et al. Whole exome sequencing reveals mutations in known retinal disease genes in 33 out of 68 Israeli families with inherited retinopathies. Sci Rep. 2015;5:13187. doi: 10.1038/srep13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel N, Aldahmesh MA, Alkuraya H, et al. Expanding the clinical, allelic, and locus heterogeneity of retinal dystrophies. Genet Med. 2016;18(6):554–62. doi: 10.1038/gim.2015.127. [DOI] [PubMed] [Google Scholar]
- 9.Tiwari A, Bahr A, Bahr L, et al. Next generation sequencing based identification of disease-associated mutations in Swiss patients with retinal dystrophies. Sci Rep. 2016;6:28755. doi: 10.1038/srep28755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao L, Wang F, Wang H, et al. Next-generation sequencing-based molecular diagnosis of 82 retinitis pigmentosa probands from Northern Ireland. Hum Genet. 2015;134(2):217–30. doi: 10.1007/s00439-014-1512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Wang H, Sun V, et al. Comprehensive molecular diagnosis of 179 Leber congenital amaurosis and juvenile retinitis pigmentosa patients by targeted next generation sequencing. J Med Genet. 2013;50(10):674–88. doi: 10.1136/jmedgenet-2013-101558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haer-Wigman L, Newman H, Leibu R, et al. Non-syndromic retinitis pigmentosa due to mutations in the mucopolysaccharidosis type IIIC gene, heparan-alpha-glucosaminide N-acetyltransferase (HGSNAT) Hum Mol Genet. 2015;24(13):3742–51. doi: 10.1093/hmg/ddv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohl S, Zobor D, Chiang WC, et al. Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia. Nat Genet. 2015;47(7):757–65. doi: 10.1038/ng.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishiguchi KM, Avila-Fernandez A, van Huet RA, et al. Exome sequencing extends the phenotypic spectrum for ABHD12 mutations: from syndromic to nonsyndromic retinal degeneration. Ophthalmology. 2014;121(8):1620–7. doi: 10.1016/j.ophtha.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Roosing S, Lamers IJ, de Vrieze E, et al. Disruption of the basal body protein POC1B results in autosomal-recessive cone-rod dystrophy. Am J Hum Genet. 2014;95(2):131–42. doi: 10.1016/j.ajhg.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Todd SC, Doctor VS, Levy S. Sequences and expression of six new members of the tetraspanin/TM4SF family. Biochim Biophys Acta. 1998;1399(1):101–4. doi: 10.1016/s0167-4781(98)00087-6. [DOI] [PubMed] [Google Scholar]
- 17.Tonisson N, Kurg A, Kaasik K, et al. Unravelling genetic data by arrayed primer extension. Clin Chem Lab Med. 2000;38(2):165–70. doi: 10.1515/CCLM.2000.025. [DOI] [PubMed] [Google Scholar]
- 18.Roberts L, Ratnapriya R, du Plessis M, et al. Molecular diagnosis of inherited retinal diseases in indigenous African populations by whole-exome sequencing. Invest Ophthalmol Vis Sci. 2016;57(14):6374–81. doi: 10.1167/iovs.16-19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YO, Oh IU, Park HS, et al. Characterization of a cDNA clone for human NAD(+)-specific isocitrate dehydrogenase alpha-subunit and structural comparison with its isoenzymes from different species. Biochem J. 1995;308(Pt 1):63–8. doi: 10.1042/bj3080063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fattal-Valevski A, Eliyahu H, Fraenkel ND, et al. Homozygous mutation, p.Pro304His, in IDH3A, encoding isocitrate dehydrogenase subunit is associated with severe encephalopathy in infancy. Neurogenetics. 2017;18(1):57–61. doi: 10.1007/s10048-016-0507-z. [DOI] [PubMed] [Google Scholar]
- 21.Rivolta C, Sweklo EA, Berson EL, Dryja TP. Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet. 2000;66(6):1975–8. doi: 10.1086/302926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Estrada-Cuzcano A, Koenekoop RK, Senechal A, et al. BBS1 mutations in a wide spectrum of phenotypes ranging from nonsyndromic retinitis pigmentosa to Bardet-Biedl syndrome. Arch Ophthalmol. 2012;130(11):1425–32. doi: 10.1001/archophthalmol.2012.2434. [DOI] [PubMed] [Google Scholar]
- 23.Riazuddin SA, Iqbal M, Wang Y, et al. A splice-site mutation in a retina-specific exon of BBS8 causes nonsyndromic retinitis pigmentosa. Am J Hum Genet. 2010;86(5):805–12. doi: 10.1016/j.ajhg.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shevach E, Ali M, Mizrahi-Meissonnier L, et al. Association between missense mutations in the BBS2 gene and nonsyndromic retinitis pigmentosa. JAMA Ophthalmol. 2015;133(3):312–8. doi: 10.1001/jamaophthalmol.2014.5251. [DOI] [PubMed] [Google Scholar]
- 25.Potter PK, Bowl MR, Jeyarajan P, et al. Novel gene function revealed by mouse mutagenesis screens for models of age-related disease. Nat Commun. 2016;7:12444. doi: 10.1038/ncomms12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartong DT, Dange M, McGee TL, et al. Insights from retinitis pigmentosa into the roles of isocitrate dehydrogenases in the Krebs cycle. Nat Genet. 2008;40(10):1230–4. doi: 10.1038/ng.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clausen W. Typisches, beiderseitiges hereditares MakulaKolobom. Klinische Monatsbldtter fur Augenheilkunde. 1921;67(116) [Google Scholar]
- 28.Koenekoop RK, Wang H, Majewski J, et al. Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nat Genet. 2012;44(9):1035–9. doi: 10.1038/ng.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiang PW, Wang J, Chen Y, et al. Exome sequencing identifies NMNAT1 mutations as a cause of Leber congenital amaurosis. Nat Genet. 2012;44(9):972–4. doi: 10.1038/ng.2370. [DOI] [PubMed] [Google Scholar]
- 30.Perrault I, Hanein S, Zanlonghi X, et al. Mutations in NMNAT1 cause Leber congenital amaurosis with early-onset severe macular and optic atrophy. Nat Genet. 2012;44(9):975–7. doi: 10.1038/ng.2357. [DOI] [PubMed] [Google Scholar]
- 31.Falk MJ, Zhang Q, Nakamaru-Ogiso E, et al. NMNAT1 mutations cause Leber congenital amaurosis. Nat Genet. 2012;44(9):1040–5. doi: 10.1038/ng.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ajmal M, Khan MI, Neveling K, et al. A missense mutation in the splicing factor gene DHX38 is associated with early-onset retinitis pigmentosa with macular coloboma. J Med Genet. 2014;51(7):444–8. doi: 10.1136/jmedgenet-2014-102316. [DOI] [PubMed] [Google Scholar]
- 33.Kiernan DF, Shah RJ, Hariprasad SM, et al. Thirty-Year follow-up of an African American family with macular dystrophy of the retina, locus 1 (North Carolina macular dystrophy) Ophthalmology. 2011;118(7):1435–43. doi: 10.1016/j.ophtha.2010.10.041. [DOI] [PubMed] [Google Scholar]
- 34.Small KW, Puech B, Mullen L, Yelchits S. North Carolina macular dystrophy phenotype in France maps to the MCDR1 locus. Mol Vis. 1997;3:1. [PubMed] [Google Scholar]
- 35.Greenwald SH, Charette JR, Staniszewska M, et al. Mouse models of NMNAT1-Leber Congenital Amaurosis (LCA9) recapitulate key features of the human disease. Am J Pathol. 2016;186(7):1925–38. doi: 10.1016/j.ajpath.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]