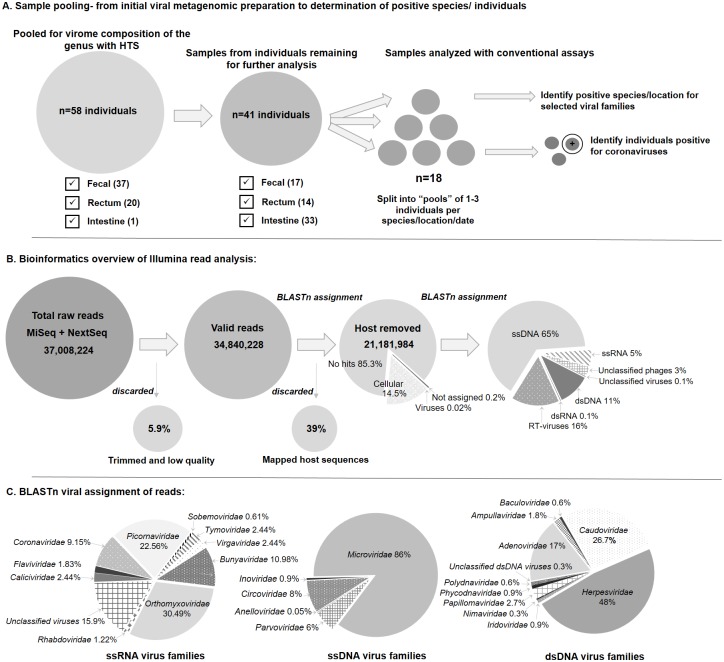
Fig 2. Strategies for sample pooling and data analysis workflows.
A) The workflow depicts the sample pooling strategies utilized in the study. Faecal and rectal samples were primarily pooled for initial viral metagenomic sample processing. Subsequently, remaining sample material (faecal homogenates, rectum, or intestine) available for individual bats were pooled according to species and location to associate selected viral families with specific species. A further step was implemented to determine specific host individuals harbouring coronaviruses in order to clarify exact sampling locations. B) Bioinformatics data analysis of Illumina reads with a developed workflow in CLC genomic workbench that trims data for quality, removes adaptors and depletes host reads based on set mapping parameters. Remaining reads were taxonomically assigned with BLASTn and MEGAN v6. C) Shows the BLASTn assignments of reads according to virus genome types: ssRNA, ssDNA and dsDNA viruses.