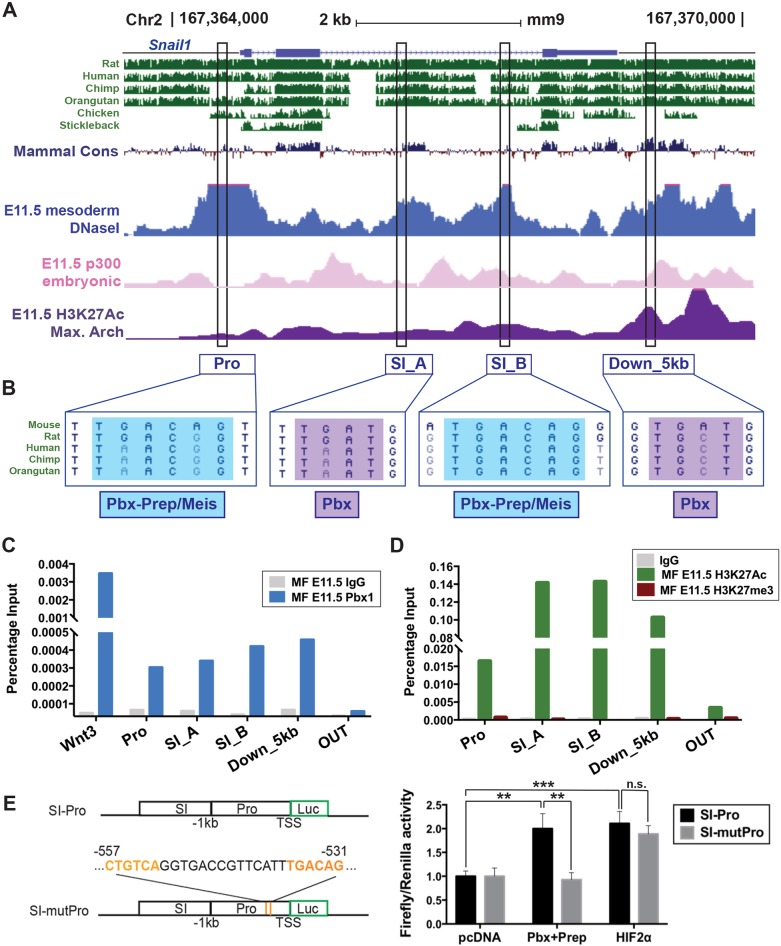
Fig. 4.
Pbx1 regulates Snail1 by binding to conserved noncoding sequences in embryonic midfacial prominences. (A) Mouse Snail1 on chromosome 2 (chr2; mm9). Peaks denote conservation across vertebrates (green); open chromatin (DNAseI hypersensitivity; blue) in mouse mesoderm (Yue et al., 2014); p300-bound sites in mouse embryonic face (pink) (Attanasio et al., 2013); and H3K27ac enrichment in maxillary arch (purple) (Landin Malt et al., 2014). Black rectangles comprise conserved noncoding sequences within Snail1 promoter (Pro), second intron (SI_A and SI_B), and downstream region (Down_5 kb) containing putative Pbx-binding sites. (B) Both Pro and SI_B encompass hexameric consensus Pbx–Prep/Meis-binding motif (TGACAG, blue); SI_A and Down_5 kb contain a partial Pbx-binding site (TGAT, purple). (C) Pbx1 binding to Snail1 in E11.5 midfaces (MF) by ChIP-qPCR. The positive control is a Wnt3 enhancer (Ferretti et al., 2011). OUT, negative control. (D) H3K27Ac and H3K27me3 ChIP-qPCR enrichment at the Snail1 locus. (E) Left top panel: mouse wild-type Snail1 SI_A (SI) and minimal promoter (Pro) cloned into pGL4 Luc reporter vector (SI-Pro). Bottom: mutant pGL4 construct (SI-mutPro) containing a deletion encompassing two Pbx–Prep-binding sites (orange) within the Snail1 promoter (position −557 to −531). Transcription start site (TSS); position 0. Right panel: Pbx-dependent transcription reporter assay in HEK293T cells. Data represented as mean±s.e.m. and calculated as firefly (Renilla) signal ratios normalized to SI-Pro activity upon transfection with empty pcDNA vector (set to 1). Wild-type Snail1 SI-Pro transactivated upon Pbx+Prep overexpression (**P<0.01). SI-mutPro reduces transactivation activity to basal levels (**P<0.01). HIF2α oxygen-insensitive form (HIF2α) transactivates Snail1 SI-Pro (***P<0.001). SI-mutPro does not affect HIF2α transactivation activity. n.s., not statistically significant.