Figure 6. Connectome of DnB to premotor and nociceptive interneuron outputs.
(A) First instar larval CNS showing reconstruction of DnB neurons (green), and nociceptive integrating interneurons (purple). Output synapses are indicated in red and input synapses in cyan. Nociceptive interneurons primarily receive input from output sites on DnB dendrites. (B) Percent of top hits’ (>3 synapses) output from right (DnB a1R) and left (DnB a1L) A1 DnB neurons as a function of cell type. Premotor circuits and nociceptive integrators are dominant outputs of DnB neurons. (C) Identities of nociceptive integrating targets for right and left DnB neurons in A1. Numbers of synapses reconstructed are indicated. Width of arrow corresponds to degree of synaptic connectivity. (D) First instar larval CNS showing reconstruction of DnB neurons (green), premotor (purple), and motor targets (yellow). Premotor neuron output synapses (red dots) located primarily in motor domain (arrowhead). (E) Identities of premotor targets for right and left DnB neurons in A1. Numbers of synapses reconstructed are indicated. Dominant outputs are A27k and A01d-3 premotor neurons. Width of arrow corresponds to degree of synaptic connectivity. (See also Figure 6—figure supplement 1).
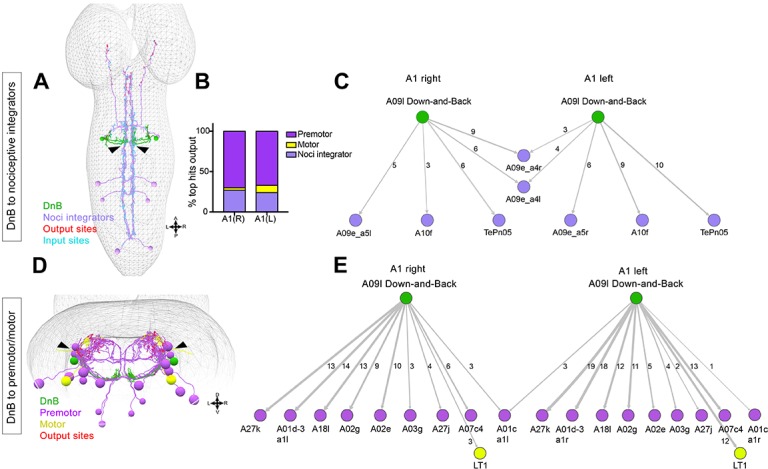
Figure 6—figure supplement 1. Additional properties of Down-and-Back output signaling.