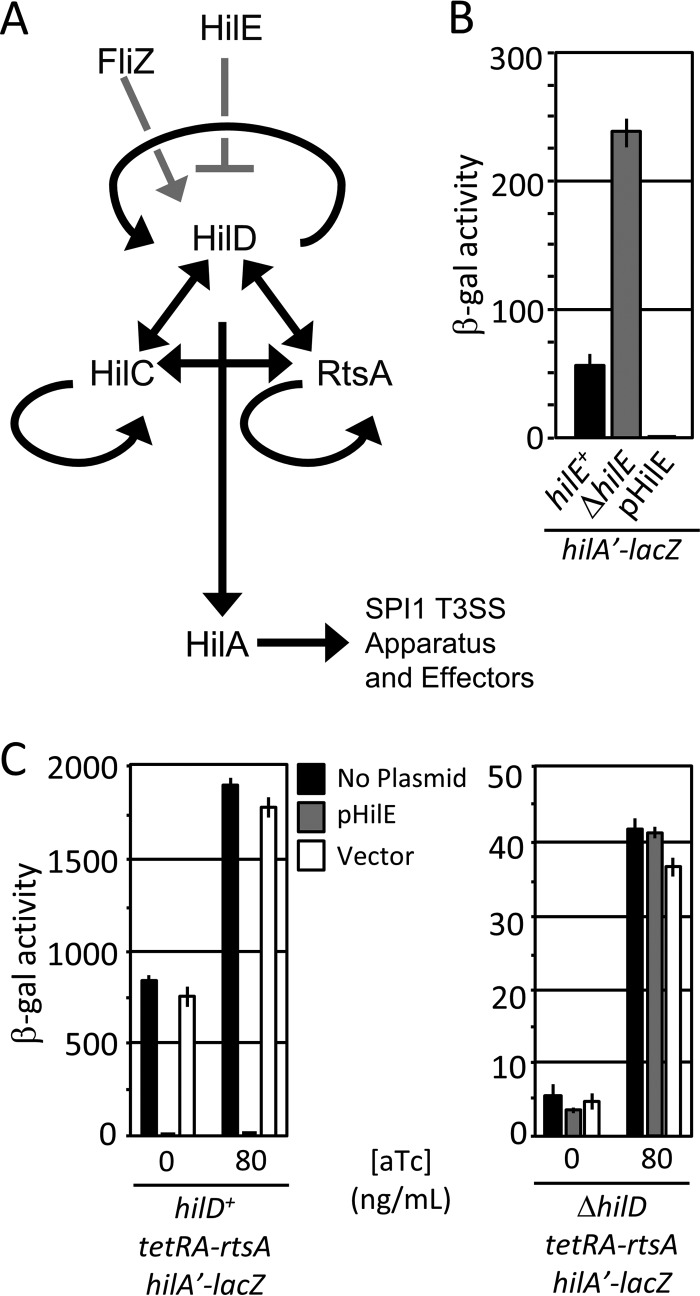
FIG 1.
(A) A simplified model for SPI1-T3SS regulation. Black lines indicate transcriptional regulation. Gray lines indicate posttranslational regulation. For clarity, the genes encoding SPI1 regulators are not shown. (B) HilE is a negative regulator of SPI1. Strains with hilE deleted or overexpressing hilE were grown under SPI1-inducing conditions and analyzed via β-galactosidase assay. β-Galactosidase activity units are defined as (micromoles of ONP formed per minute × 106)/(OD600 × milliliters of cell suspension) and reported as mean ± one standard deviation (n = 6, 3 sets of duplicate assays on 3 separate days). The strains used were JS749, JS749 with pHilE, and JS2103. (C) HilE acts via HilD. The rtsA gene was under the control of a tetA promoter, hilD was left unaltered or deleted, and HilE was overexpressed as indicated. Strains were grown overnight under SPI1-inducing conditions and subcultured 1:100 with the indicated concentration of anhydrotetracycline (aTc) (n = 4, 3 sets of duplicate assays on 2 separate days; error bars represent one standard deviation). The strains used were JS953 and JS2104 with pHilE or pWKS30.