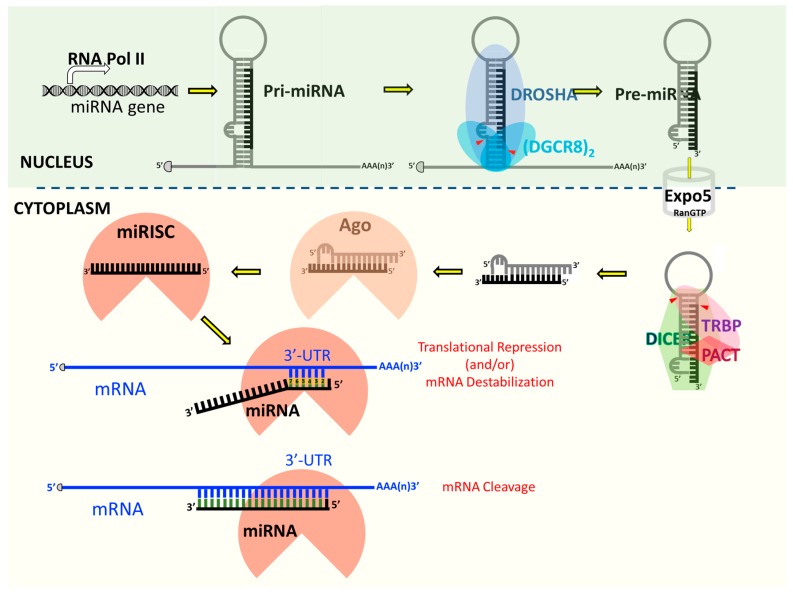
Figure 1.
Schematic representation of the canonical microRNA (miRNA) biogenesis pathway in humans: The RNA Polymerase II transcribes the nuclear chromosome-encoded miRNA gene into primary miRNA (pri-miRNA) that comprises a stem, a terminal loop, and single-stranded 5′ and 3′ RNA tails. The nuclear microprocessor complex-resident Drosha, in concert with its cofactor DGCR8, recognizes the pri-miRNA and cleaves asymmetrically at defined sites in the stem. The released stem-loop structure termed precursor miRNA (pre-miRNA) is then transported to the cytoplasm by the Exportin 5-RanGTP protein complex. The cytoplasmic Dicer, in concert with potential cofactors like transactivating response RNA-binding protein (TRBP) and protein activator of the interferon-induced protein kinase (PACT), recognizes the 2-nucleotide 3′ overhang and/or the 5′ phosphorylated end of the pre-miRNA and cleaves at a defined distance from those termini. The ensuing asymmetric ~22-nucleotides-long miRNA duplex, marked by the 5′ monophosphate and a 2-nt 3′ overhang on both strands, is then loaded onto the AGO protein complex in an ATP-dependent process aided by chaperones and co-chaperones. The miRNA duplexes are then unwound and one of the strands (passenger strand), selected on the basis of 5′ nucleotide composition and thermodynamic stability, concomitantly undergoes dissociation or degradation. The resultant mature miRNA (guide strand), in the context of the effector nucleoprotein complex called miRNA-induced silencing complex (miRISC), then binds the target mRNA leading to translational suppression or mRNA deadenylation or mRNA degradation.