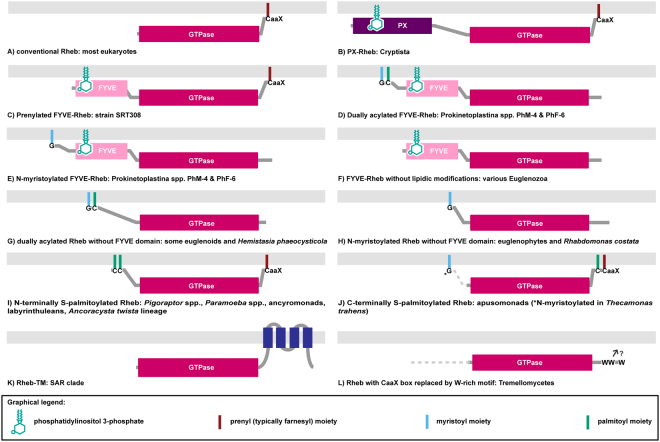
Figure 1.
The diversity of membrane-association mechanisms of Rheb proteins. (A) Conventional Rheb. (B) PX-Rheb in Cryptista (for details see Supplementary Fig. S2). (C–H) Unconventional Rheb forms in Euglenozoa and relatives (see Supplementary Fig. S3 for details). (C) Prenylated FYVE-Rheb. (D) Dually acylated (N-myristoylated and S-palmitoylated) FYVE-Rheb. (E) N-myristoylated FYVE-Rheb. (F) FYVE-Rheb without lipidic modifications. (G) Dually acylated (N-myristoylated and S-palmitoylated) Rheb without the FYVE domain. (H) N-myristoylated Rheb without the FYVE domain. (I–J) S-palmitoylated Rhebs occurring in various protists (for details see Supplementary Fig. S5). (I) N-terminally S-palmitoylated Rheb; note that the number of predicted S-palmitoylation sites vary from one to seven in this class (see Supplementary Table S4). (J) C-terminally S-palmitoylated Rheb; note that the two proteins known in this category differ in the number of S-palmitoylation sites (one or two; see Supplementary Table S4) and in the presence/absence of N-myristoylation. (K) Rheb-TM in the SAR clade (for details see Supplementary Fig. S6). (L) Rheb in the Tremellomycetes with the CaaX box replaced by a W-rich motif also possibly (question mark) mediating a membrane association (for details see Supplementary Fig. S7).