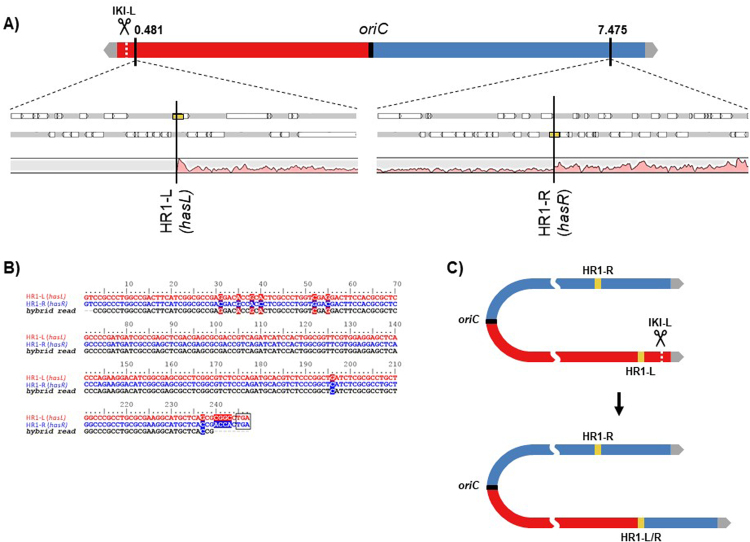
Figure 5.
Chromosomal arm replacement by homologous recombination after DSB induction at IKI-L. (A) Analysis of the coverage of sequencing reads along the chromosome of a DSB survivor at IKI-L (WT17). Genome sequencing reads of strain WT17 plotted along the S. ambofaciens ATCC 23877 genome as a reference are shown as a pink area. Yellow boxes symbolize HR1-R and HR1-L (two duplicated copies of a sigma factor encoding gene: hasL, SAM23877_0517, and hasR, SAM23877_6930). (B) Hybrid read resulting from recombination between HR1-L and HR1-R sequences. HR1-L and HR1-R sequences were aligned with one typical hybrid_read found in WT17 sequencing data. The alignment shown covers the 3’ end of the hasL/R sequences (from position 600 to 846). Polymorphism is symbolized with a colored background. The stop codon is boxed. (C) WT17 chromosome structure. The hybrid HR1 sequence is labelled HR1-L/R. The I-SceI cut at IKI-L resulted in the loss of the left chromosomal arm and in a homologous recombination event between HR1-R and HR1-L leading to the duplication of the right chromosomal arm over 825 kb.