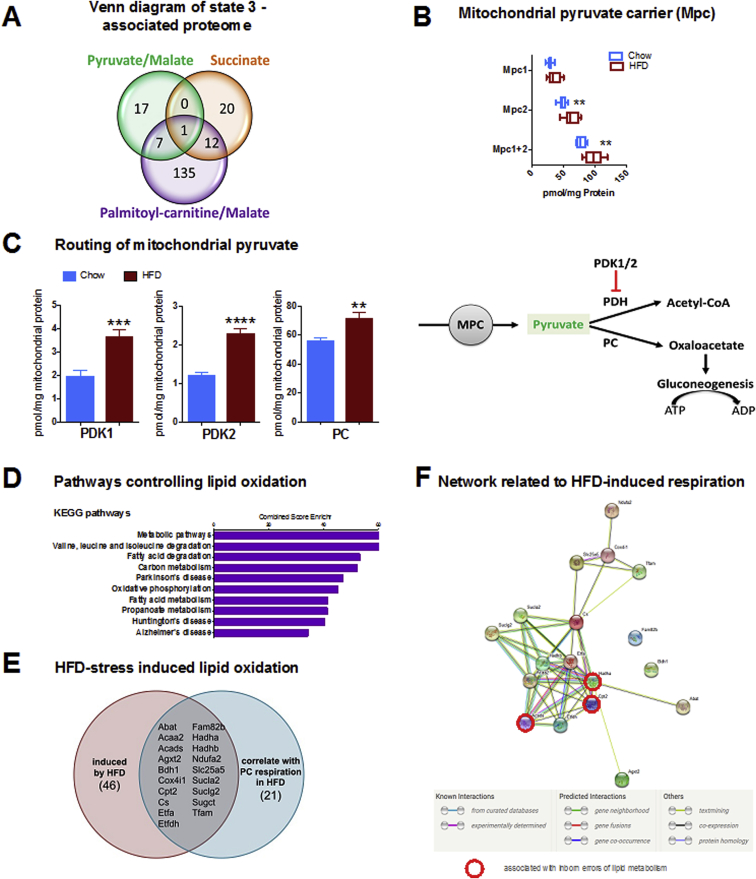
Figure 3.
Correlation analysis of mitochondrial respiration and proteome. (A) 976 mitochondrial proteins (as annotated in the Mitocarta 2.0) were correlated with state 3 respiration for different substrates. The numbers in the Venn-diagram depict significantly correlating proteins (Spearman ρ ≥ 0.5, p < 0.05). The single protein correlating with all substrates is MPC1. (B) Absolute protein concentrations of Mpc1, Mpc2, and Mpc1 + Mpc2. (C) Routing of mitochondrial pyruvate during HFD-feeding based on proteomic data. Enhanced imported pyruvate is not decarboxylated, as increased levels of pyruvate dehydrogenase kinases PDK1 and PDK2 inhibit pyruvate dehydrogenase (PDH). Increased pyruvate carboxylase (PC) levels support enhanced pyruvate carboxylation, forming oxaloacetate for gluconeogenesis, a highly ATP-demanding process driving respiration. (D) Enrichment analysis for KEGG pathways was performed for proteins significantly correlating with state 3 palmitoyl-carnitine (PC)/malate respiration. The top 10 significantly enriched KEGG pathways are displayed with their respective combined score calculated by Enrichr. (E) Prediction of proteins controlling lipid respiration in response to nutrient stress. Protein levels significantly induced by HFD (p < 0.05) were filtered specifically for correlation with HFD-induced PC/malate state 3 respiration. (F) The functional link between HFD-stress induced proteins is strongly supported by the string-db network [30]. Acads, Cpt2, Hadha (indicated by red circle) were previously identified as disease genes for inborn errors of lipid metabolism [28]. All data represent n = 8 animals. Boxplots indicate 25–75 percentiles, with the vertical line indicating the median and whiskers from minimum to maximum (B) or as mean ± standard error of mean (C). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 in a t-test comparing chow vs HFD.