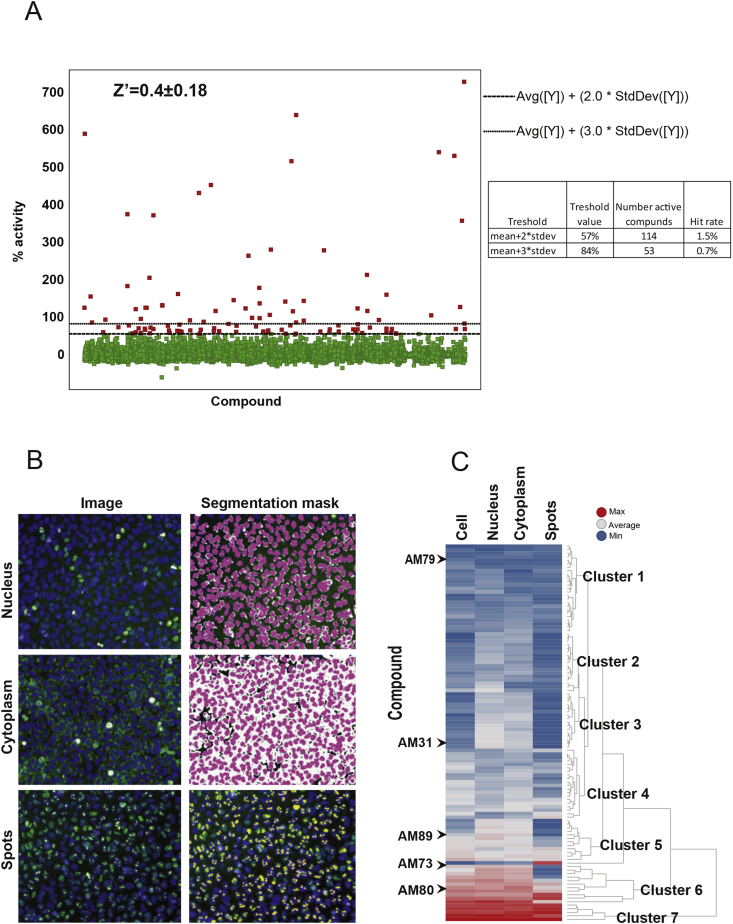
Figure 2.
Screening results and cut-off selection. (A) Distribution of EGFP-PGC-1α1 signal and selection of the cut-off value after screening of 7040 compounds (approximately 25% of the library). The mean and standard deviation of % activation (Intensity Cell Sum − Median per Well) for all tested compounds were calculated according to (1 − ((x − mean [MIN])/(mean[MAX] − mean[MIN])) * 100%), where MIN refers to the negative controls (DMSO-treated cells) and MAX refers to the positive controls (MG132-treated cells). A threshold value of mean + 2 ∗ stdev was applied to select active compounds. The table on the right panel shows the number of hits and hit rates with two different cut-offs. (B) Segmentation masks were used to identify where the EGFP-PGC-1α1 signal was located either; nucleus, cytoplasm or aggregation. Example of fluorescence picture on the left and corresponding segmentation mask to the right. (C) Clustering of hits according to similarities in cellular distribution of EGFP-PGC-1α1 signal. Clustering of hits is shown on the right side and five selected compounds chosen for further investigation are shown under their respective cluster.