Fig. 2.
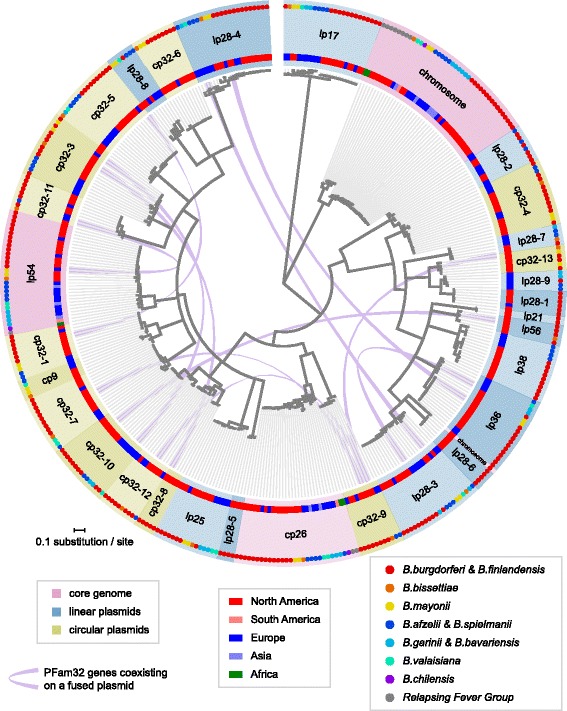
PFam32 gene tree suggests ancestral radiation of plasmid compatibility groups. The midpoint-rooted gene tree (center) is based on a 392-amino acid long alignment of 411 PFam32 homologues from 34 Borreliella genomes and selected sequences from 8 relapsing-fever Borrelia genomes. The rings, from center outward, indicate the (i) continent of origin of each isolate, (ii) plasmid compatibility type names with different shades of pink, blue, and green for the core genome (chromosome, cp26 and lp54), linear, and circular plasmids, respectively, and (iv) species (colored dots). Lavender central ribbons connect multiple PFam32 alleles found on the same replicon, indicating plasmid fusion events. A vertical PFam32 tree is available as Additional file 1: Figure S1, where two PFam32 homologs (one on lp28–9 from BOL26, another on lp56 from DN127) with positions inconsistent with the genome phylogeny (see Fig. 5 below) – indicative of whole-plasmid transfer – are highlighted. Plasmid types lp5 and some cp9s are not shown in the figure as they do not have a PFam32 partition gene
