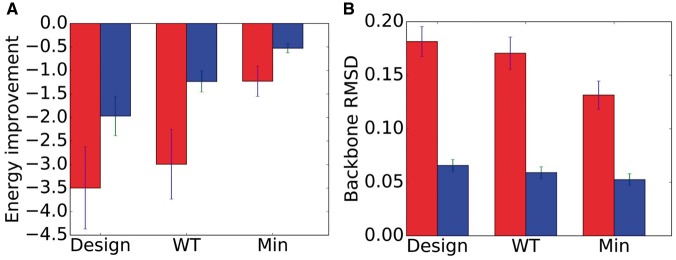
Fig. 2.
Seventy-nine computational experiments comparing CATS, DEEPer and rigid-backbone design. (A) Average improvement in energy (kcal/mol) in CATS (red) and DEEPer (blue) calculations compared to rigid-backbone calculations. Averages with standard error bars shown for designs, wild-type (WT) conformational searches, and single-voxel minimizations starting from the wild-type conformation. (B) RMSD (Å) between crystal-structure backbones and optimal backbones computed by CATS (red) and DEEPer (blue) for the same test cases as (A). CATS is able to model larger backbone changes, and the greater RMSD for designs compared to minimizations indicates CATS is modeling the backbone shifts induced by mutations