Fig. 4.
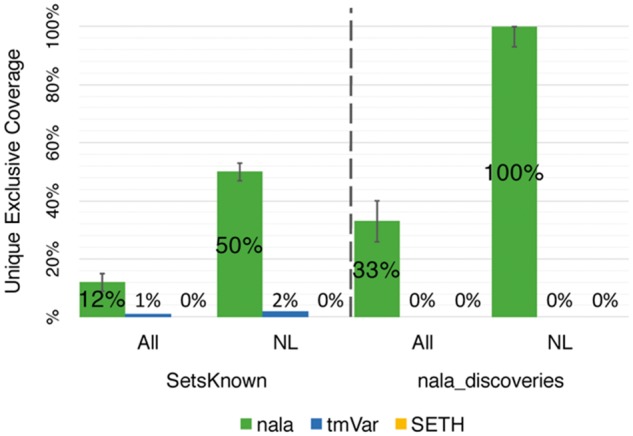
nala could fully replace other methods. For each publication we considered all mentions correctly identified by one of the top three methods and kept only the findings unique in each publication. The y-axis plots the percentage of those mentions identified uniquely by one of the methods (All: all mentions, NL: NL-only mentions). For all corpora containing publications of genes and proteins indexed in the databases (SetsKnown), 1% of the mentions were detected only by tmVar and 12% only by nala, while SETH found no mention in this dataset that nala had not detected. Only nala correctly detected NL-only mentions in abstracts with new discoveries (100% bar on right triplet)
