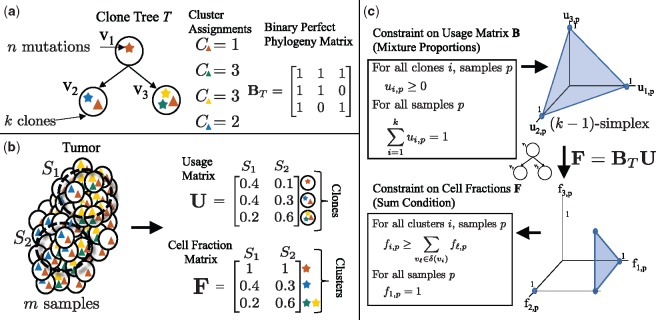
Fig. 1.
Tree constraint on cluster cell fractions. (a) We model the evolution of a tumor as a clone tree T, with k vertices corresponding to clones in the tumor. A mutation (denoted here by a star) is assigned to the clone in which it originates. Under the infinite sites assumption, a mutation occurs once, and is never lost. Thus, if a mutation occurs in clone vi, all descendent clones of vi will also contain that mutation. A clone tree T can be described by a binary perfect phylogeny matrix BT. (b) We measure m samples from a heterogeneous tumor, each sample containing a mixture of clones. The usage matrix U describes the proportion of each clone in each sample. The cell fraction matrix F describes the proportion of cells that contain a given cluster of mutations. For example, here the clone v1, containing the red mutation, occurs in of sample S1, but has a cell fraction of , as the red mutation is present in all cells. (c) As the usage matrix U describes mixture proportions, the columns of U are constrained to be on the -simplex. For a tree T, F and U are related by F = BTU. Thus, the set of allowed cell fractions for a tree T is a linear transformation of the -simplex and is unique for every distinct tree T. This set can be described by the Sum Condition, where denotes the set of children of vi in T