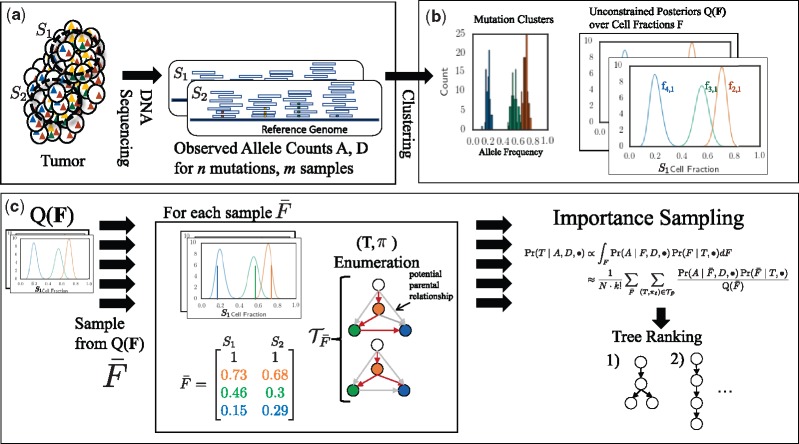
Fig. 3.
Overview of PASTRI algorithm. (a) We observe variant-allele read counts A and the total number of reads D that align to the locus for n mutations across m samples of the tumor. (b) A clustering algorithm that does not require that the data is generated by a phylogenetic tree gives an estimate of the posterior distribution over cluster cell fractions F. (c) PASTRI draws samples from . For each sample , PASTRI enumerates the set of trees T and assignments π of cell fractions to vertices of of T that satisfy the Sum Condition. All trees/vertex-assignment pairs not in have a probability of 0. Algorithm estimates the posterior probability of each tree using importance sampling