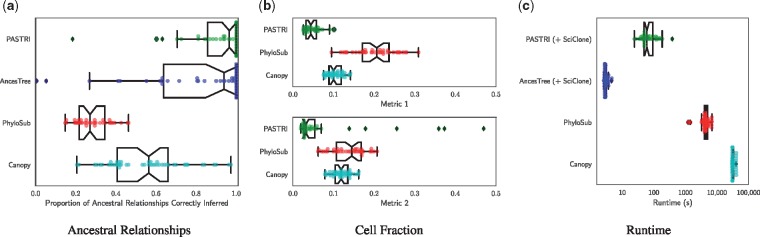
Fig. 5.
Comparison of phylogenetic reconstruction algorithms. We simulate 50 trees with 5 vertices, 5 samples and 20 mutations. (a) To quantify the accuracy of the tree inference, we measure the proportion of ancestral relationships that the algorithms correctly recover. A pair of mutations c and d has four possible ancestral relationships: c is ancestral to d, d is ancestral to c, c and d are in the same cluster, or c and d are on separate branches. (b) To measure how accurately the algorithms recover the true cell fractions, we report two metrics, given by Equations 13 and 14. Both metrics measure the average distance between the true cluster cell fraction matrix F and the reported cluster cell fractions. Metric 1 penalizes for overestimating the number of clusters, and Metric 2 penalizes for underestimating. (c) We report the runtime in seconds of each method, where the x-axis is a logarithmic scale