Abstract
The late assembly (L) domain of retrovirus Gag, required in the final steps of budding for efficient exit from the host cell, is thought to mediate its function through interaction with unknown cellular factors. Here, we report the identification of the Nedd4-like family of E3 ubiquitin protein ligases as proteins that specifically interact with the Rous sarcoma virus (RSV) L domain in vitro and in vivo. We screened a chicken embryo cDNA expression library by using a peptide derived from the RSV p2b sequence, isolating two unique partial cDNA clones. Neither clone interacted with a peptide containing mutations known to disrupt in vivo RSV L domain function or with human immunodeficiency virus type 1 (HIV-1) and equine infectious anemia virus (EIAV) L domain-derived peptides. The WW domain region of one of the clones, late domain-interacting protein 1 (LDI-1), but not the C2 domain, bound RSV Gag and inhibited RSV Gag budding from human 293 cells in a dominant-negative manner, functionally implicating LDI-1 in RSV particle budding from cells. RSV Gag can be coimmune precipitated from cell extracts with an antisera directed at an exogenously expressed hemagglutinin (HA)-tagged LDI-1 or endogenous Nedd4 proteins. These findings mechanistically link the cellular ubiquitination pathway to retrovirus budding.
Keywords: RSV, Gag, virus particle assembly
One of the least understood aspects of viral replication is the assembly of virions and their exit from the host cell. However, it has been demonstrated that determinants of retroviral assembly and budding lie solely within the Gag polyprotein (1, 2). Mutational analyses of Gag revealed three distinct domains involved in virion assembly: a membrane-binding domain (M domain), found at the amino terminus of matrix (MA), which contains a myristoylation signal in most retroviruses (1, 3, 4); an internal Gag interaction domain (I domain), which maps to nucleocapsid (NC) in RSV (5), and is involved in the aggregation of Gag polyproteins necessary for particle formation; and a late assembly domain (L domain), required late in the budding process (6–10).
For Gag particle budding, a functional L domain is not required on every Gag molecule (9); moreover, the L domain can function at other positions in Gag, suggesting that it is a protein interaction domain (10). The RSV Gag L domain maps specifically to a proline-rich sequence of p2b, PPPPYV, located between the MA and capsid (CA) proteins in Gag (9, 10). Referred to as the PY motif, this sequence is found in the Gag protein of many retroviruses, but not lentiviruses, and in numerous cellular proteins. Interestingly, this motif is also found in the structural proteins of other budding viruses, including rhabdoviruses (vesicular stomatitis virus) and filoviruses (Ebola and Marburg viruses; ref. 11). For VSV, this PY motif has recently been shown to function in the late steps of budding, analogous to the L domain of the retroviral Gag (12). The PY motif specifically binds to a protein-interaction domain found mostly in signaling and regulatory proteins termed the WW domain. It is ≈40 aa in length and contains 4 conserved aromatic amino acids of which 2 are invariant tryptophan residues (13–16).
A proline-rich L domain equivalent maps to a PTAP sequence in the N-terminal region of HIV-1 p6, a region at the C terminus of Gag (6, 7) and a non-proline-rich equivalent maps to a YPDL sequence (the YXXL motif) in the analogous p9 region of EIAV Gag (8, 17). The p6 region of HIV-1 Gag was originally suspected to be involved in virus assembly because frame-shift mutations introduced into this region prevented the release of tethered viral particles from cells (18). Both lentivirus L domains are interchangeable with the RSV PY motif despite a lack of sequence identity among the three sequences (8, 17). Puffer et al. reported that the equine infectious anemia virus (EIAV) L domain interacts with the AP-50 subunit of the cellular AP-2 clathrin-associated adaptor protein complex (19). This interaction was specific to EIAV p9, because HIV-1 p6 and Rous sarcoma virus (RSV) p2b late domains did not bind to this protein.
The known properties for the three different retroviral L domains suggest that they are protein-interaction domains, and most likely function by binding to specific cellular proteins that facilitate the late stages of retroviral particle release. The differences in sequences of the three L domains suggest either of two possibilities: (i) each L domain uses an independent budding pathway to exit the cell; or (ii) the three L domains bind to different components of the same budding pathway to exit the cell. To test these hypotheses, we screened a chicken cDNA expression library, by using a peptide derived from the RSV L domain, for cellular factors involved in L domain function. Two unique clones [named late domain-interacting (LDI) protein 1 and 2] encoding members of the neuronal precursor cell-expressed developmentally down-regulated 4 (Nedd4) family of E3 protein ubiquitin ligases were isolated. The binding between each of these two proteins and the RSV L domain peptide in vitro appeared to be specific because neither a mutant RSV L domain peptide nor wild-type HIV-1 or EIAV L domain-derived peptides bound either of the proteins. When cotransfected with RSV Gag, a fragment of LDI-1 that contains the four WW domains inhibited Gag budding from human 293 cells in a dominant-negative manner. These data, together with coimmune precipitation data, suggest that we identified a cellular factor, LDI-1, an E3 ubiquitin protein ligase, which mediated RSV L domain function.
Materials and Methods
Antibodies.
The anti-influenza hemagglutinin (HA) epitope rabbit polyclonal and mouse monoclonal (HA.11) antibodies were purchased from Covance (Richmond, VA). The anti-RSV rabbit polyclonal antibody was a gift from John Wills, Penn State University (9, 10). The RSV p19 mouse monoclonal antibody, C3A, was purchased from the University of Iowa Hybridoma Bank. Rabbit anti-rat Nedd4 polyclonal antibody was a gift from Daniela Rotin, Hospital for Sick Children, Toronto, Canada (20). Rabbit anti-mouse Nedd4 polyclonal antibody was a gift of Sharad Kumar, Institute of Medical and Veterinary Science, Adelaide, Australia (21). Preadsorbed alkaline phosphatase-conjugated rabbit and mouse secondary antibodies were from Santa Cruz Laboratories (Santa Cruz, CA), whereas 35S-labeled donkey anti-rabbit and sheep anti-mouse secondary sera were from Amersham Pharmacia.
Plasmids Constructs.
Plasmid 2036, a derivative of 1920 (22) that contains the SV40 origin/early promoter, a G418-resitance marker, a cytomegalovirus-enhanced green fluorescent protein (CMV-EGFP) cassette, and the Epstein–Barr virus (EBV) FR plasmid maintenance element, was used as the backbone for the plasmid constructions for expression in human cells. The gag gene from the Prague C strain of RSV, bearing a protease-inactivating mutation (D37S), was subcloned from pMyr0 (9) into the 2036 plasmid by using KpnI and XbaI, replacing EGFP with gag. LDI-1 and LDI-1 derivatives, within the 2036 backbone, were N-terminally tagged with the HA epitope YPYDVPDYA. KpnI and XbaI restriction sites were introduced by standard PCR into all of the LDI-1 constructs at 5′ and 3′ ends, respectively, and used for subcloning into plasmid 2036. For details of the primer sequences used in the PCR of the various LDI fragments, see Materials and Methods and figs. 6–9, which are published on the PNAS web site, www.pnas.org. AGP5 is an SV40 large T antigen expression vector derived from 1606 (23) that contains a functional SV40 origin. The pRSVL (RSV-luciferase) was previously described (22). The RSV Gag-EGFP plasmid was a gift from John Wills, Pennsylvania State University. pcDNA3 was used as filler DNA and was from Invitrogen.
Screening a Chicken Embryo cDNA Phage Expression Library.
Plaque forming units (pfu) (1 × 106) of a chicken embryo cDNA expression library (CLONTECH), constructed in λgt11, were screened by using standard protocols (24). Filters were probed with 3 mM biotinylated-peptide probe, prebound to alkaline phosphatase-conjugated NeutrAvidin (Pierce). A standard alkaline phosphatase colorimetric assay was used to detect hybridized probe. Lambda DNA was harvested from purified positive plaques (24), and excised cDNA inserts subcloned into the EcoRI site of pGEM 3zf(−) plasmid (Promega). Sequencing was carried out by using an Applied Biosystems model 377 automated sequencer. For details of the screen, including sequences of the biotinylated RSV, HIV-1, and EIAV L domain peptides used, refer to supporting information, which is published on the the PNAS web site, www.pnas.org.
Transfection and Metabolic Labeling of Cells.
All cell lines were maintained in DMEM-high glucose media supplemented with 10% FBS, 1,000 units/ml penicillin, and 1,000 μg/ml streptomycin, unless otherwise specified. DNAs were introduced into cells by calcium phosphate transfection (24). The human 293/E cell line used in this study is a derivative of the adenovirus transformed human embryonic kidney cell line 293 (25) that stably expresses the EBNA1 protein of EBV. Cells were plated at 40% confluence 24 to 30 h before transfection. Each 10-cm plate was transfected with 20 μg of total DNA. Unless otherwise specified, each transfection included 5 μg of the RSV Gag expression plasmid, 100 ng each of AGP5 and pRSVL, varying amounts of DNA encoding the HA-tagged proteins (LDI-1, LDI-2, LDI-1 WW domain, or LDI-1 C2 domain; 1.5–15 μg), and pcDNA3 as filler DNA. pRSVL was included in all transfections to correct for transfection efficiency by measuring luciferase expression. Luciferase expression was assayed by using the Luciferase Assay System kit (Promega) as per the manufacturer's instructions. Cells were metabolically labeled as previously described (9, 10). For details, see supporting information.
Coimmune Precipitation of Proteins from Cells.
COS-1 cells (3 × 106) were transfected with 10 μg of RSV Gag-GFP plasmid by using Fugene 6 transfection reagent (Roche) and harvested 48 h post transfection. Cells were washed three times with cold PBS, swollen in 1 ml hypotonic buffer (10 mM Tris⋅HCl, pH 7.4/1 mM MgCl2), and lysed with 30 strokes in a Dounce homogenizer with a type B pestle. Cell extracts were subjected to centrifugation at 1,000 × g for 10 min at 4°C to sediment nuclei, followed by centrifugation at 10,000 × g for 10 min at 4°C to pellet organelles and cytoskeleton components. The soluble fraction obtained (S10) was analyzed by immunoprecipitation followed by SDS/PAGE and Western blotting (26), using antibodies described in the text.
Results
Identification of Cellular Proteins That Interact with the RSV Gag L Domain.
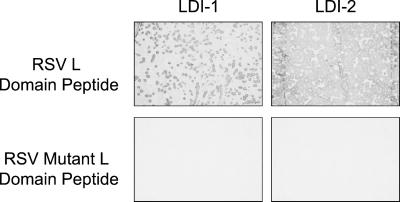
A peptide comprised of 3 tandem copies of the 11-aa p2b sequence, which contains the L domain, was used to screen a chicken cDNA expression library. Two unique cDNA expression clones with inserts of ≈1.7–1.8 Kb were identified by our screening protocol. To determine whether binding to the RSV L-domain probe was specific, the two cDNA expression clones were screened by using monomeric or trimeric peptide probes in which the PY motif was mutated by amino acid substitutions previously shown to disrupt L domain function in vivo (10). The encoded protein products from either clone did not bind to the mutant RSV L domain peptides (Fig. 1). Nor did the encoded protein products from the two clones interact with core trimeric HIV-1 p6 or EIAV p9 L domain peptide probes (data not shown). These results demonstrated that the interactions observed in vitro were specific for the wild-type RSV L domain sequence and were mediated by the PY motif.
Figure 1.
Chicken embryo cDNA clones express proteins that specifically interact with the RSV L domain PY motif in vitro. Individual phage clones from a chicken embryo cDNA expression library were screened for protein products that interact with the indicated biotinylated L domain probe (as described in Materials and Methods). Purified phage are as indicated. Positive controls used a plasmid expressing a YAP WW-GST fusion protein. A λgt11 cDNA clone that did not react with the RSV wild-type trimeric probe was used as a negative control. In this figure, the RSV and RSV mutant refer to RSV trimeric wild-type and mutant peptide probes, respectively.
The two clones have been named LDI-1 and -2. A blast analysis of nucleotide and protein sequence databases matched both clones to several WW domain-containing proteins. The best matches were with the Nedd4-like family of E3 ubiquitin protein ligases. The sequences and relationship to Nedd4-like E3 proteins are described in the supporting information. Both cDNAs encoded partial E3 protein sequences lacking the C-terminal HECT domain. On expression, both proteins crossreacted with anti-rat Nedd4 WW2 and anti-mouse Nedd4 polyclonal antibodies, confirming that they are members of this family of proteins (data not shown).
Coimmune Precipitation of Gag and Nedd4-Related Protein(s) from Cos-1 Cells.
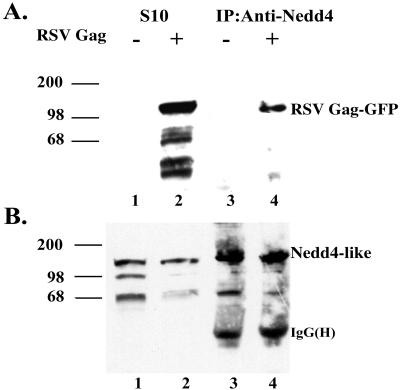
To determine whether Gag interacted with Nedd4-related proteins in vivo, we attempted to coimmune precipitate Gag and the endogenous Nedd4-related proteins from extracts of COS-1 cells transfected with a RSV Gag-EGFP expression plasmid. For the coimmune precipitation, we used a rabbit anti-rat Nedd4 WW2 sera and analyzed precipitated complexes by immunoblotting using either anti-RSV p19Gag mouse monoclonal antibody or anti-rat Nedd4 WW2 sera as shown in Fig. 2. No Gag-related protein was recognized in untransfected cells (Fig. 2A, lane 1) whereas a protein of ≈110 kDa corresponding to the RSV Gag-EGFP fusion protein was detected in the transfected cells (lane 2). After immune precipitation with the anti-rat Nedd4 WW2 sera, the RSV Gag-EGFP fusion protein was detected in precipitates from transfected cells (lane 4), but not control cells (lane 3). As a control, the membranes were reprobed with the anti-rat Nedd4 WW2 sera. Bands of ≈130, 100, and 70 kDa were detected in the S10 fractions and immune precipitated samples (Fig. 2B, lanes 1–4). The 130-kDa band was enriched in the immune precipitated samples and exhibited the molecular weight and immune crossreactivity of full-length Nedd4 (21), suggesting it is a member of the Nedd4 family. We conclude from these data that Nedd4-related protein(s) interact with RSV Gag in COS-1 cells.
Figure 2.
Coimmune precipitation of Gag and Nedd-4 like protein from COS-1 cells. (A) Untransfected COS-1 cells (lanes 1 and 3) or cells transfected with pCMV-RSVGag-EGFP (lanes 2 and 4) were lysed and processed to prepare S10 extracts as described in Materials and Methods. Lanes 1 and 2 represent the input S10 extracts, whereas lanes 3 and 4 represent the S10 extracts subjected to immune precipitation with anti-Nedd4 sera. Proteins were resolved by SDS/PAGE and detected by Western blotting with an RSVGag-p19 monoclonal antibody. (B) The immunoblot described in A was reprobed by using an anti-Nedd WW domain antibody.
L Domain Required for Budding of RSV Gag from 293 Cells.
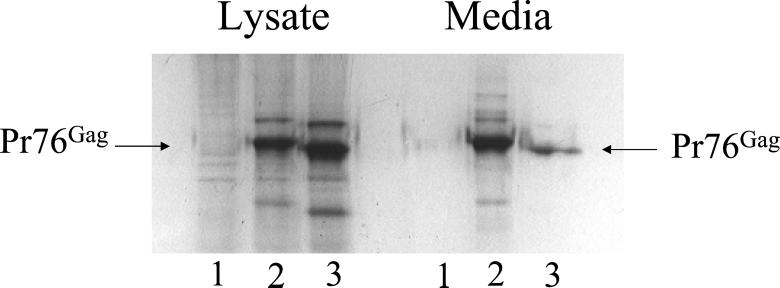
We chose to test the role of the LDI proteins in RSV Gag budding by using human 293/E cells. These cells constitutively express the EBV EBNA1 protein. We first demonstrated that budding of RSV Gag from these cells required an L domain by transfecting them with a Gag L domain deletion construct (Δp2b), previously analyzed in COS-1 cells (9, 10). To do these experiments, we placed an EBV FR maintenance element and the transcriptional enhancer of EBV into our Gag expression plasmid. These elements result in a high steady state level of Gag expression in the presence of the EBNA1 protein in the 293/E cells. As shown in Fig. 3, virus-like particles are released from the 293/E cells into the media, similar to what was observed when RSV Gag was transiently expressed in COS-1 cells. Because the Gag expression plasmid used in these studies had a PR inactivating-D37S mutation, we detected only full-length Pr76Gag and not processed proteins. When the RSV Gag Δp2b/D37S mutant was substituted for wild-type Gag, there was a greater than 90% reduction in Gag released into the media, confirming that the L domain is required for efficient budding from 293/E cells (Fig. 3, lane 3).
Figure 3.
The RSV L domain is required for Gag to bud from 293/E cells. Cells were transfected with a plasmid encoding wild-type or L domain-deleted (Δp2b) Gag as described in Materials and Methods. [35S]methionine-labeled cell lysates and media were immune precipitated with an anti-RSV rabbit polyclonal antiserum, and the proteins were fractionated by SDS/PAGE. The amount of Pr76Gag in the lysate and media fractions was determined by phosphorimage analysis. Mock-transfected cells (lanes 1), wild-type Gag (lanes 2), and Δp2b Gag (lanes 3).
The WW Domains of LDI-1 Act as a Dominant-Negative Inhibitor of RSV Gag Budding from Human 293 Cells.
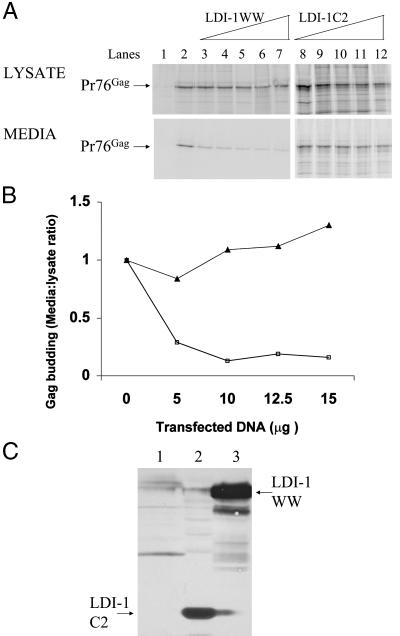
To test whether there is a biologically relevant interaction between the LDI-1 protein and RSV Gag in vivo, we tested whether a high level of expression of LDI-1 interfered with the ability of the endogenous Nedd4-related protein(s) to participate in Gag budding. Preliminary experiments conducted by cotransfecting varying amounts of HA-tagged LDI-1 expression plasmid with a fixed concentration of RSV Gag expression plasmids suggested that expression of LDI-1 dominantly interfered with Gag budding (data not shown). To define the region of LDI-1 responsible for this dominant-negative inhibition of budding, we tested the HA-tagged C2 (amino acids 1–131) and WW domains (amino acids 132–571) of LDI-1. As shown in Fig. 4A (lanes 3–7), the cotransfection of an expression plasmid encoding the LDI-1 WW domains inhibited RSV Gag budding to greater than 90%. In contrast, the cotransfection of the LDI-1 C2 domain expression plasmid (lanes 8–12) did not interfere with RSV budding. Gag protein containing bands were quantified by phosphorimage analysis using a Molecular Dynamics PhosphorImager, and the level of Gag budding was represented as the ratio of Pr76Gag in the media to the lysate (Fig. 4B). Immunoblot analysis performed with a rabbit anti-HA polyclonal antiserum confirmed expression of the HA-tagged proteins (Fig. 4C). Similar results were obtained by using COS-1 cells transfected with the RSV-EGFP construct, indicating that the dominant negative effect of the LDI-1 WW domains is cell independent (data not shown). In a control experiment, cotransfection of an expression plasmid encoding β-galactosidase did not decrease budding of Gag from cells (data not shown). These results suggest that the WW domains of LDI-1 mediate the dominant-negative interference of RSV Gag budding. Moreover, preliminary studies indicate that the individual WW domains differ in their ability to mediate this inhibition (data not shown), indicating that the effect is specific.
Figure 4.
Dominant-negative inhibition of RSV budding in 293/E cells by the LDI-1 WW domains. (A) Cells were transfected with the wild-type RSV Gag expression plasmid along with increasing amounts of either an expression plasmid for the WW domains of LDI-1 (lanes 3–7), or the LDI-1 C2 domain (lanes 8–12). Transfected cells were labeled with [35S]methionine, and Gag was immunoprecipitated from cell lysates and media, separated by SDS/PAGE, and quantified by phosphorimage analysis. Untransfected cells (lane 1), cells transfected with wild-type Gag (lane 2). Cells transfected with Gag and varying amounts of LDI-1 WW as follows: 5 μg (lane 3), 7.5 μg (lane 4), 10 μg (lane 5), 12.5 μg (lane 6), and 15 μg (lane 7). Cells transfected with Gag and varying amounts of LDI-1 C2 as follows: 5 μg (lane 8), 7.5 μg (lane 9), 10 μg (lane 10, 12.5 μg (lane 11), and 15 μg (lane 12). (B) The effect of cotransfection of plasmids expressing the WW domains of LDI-1 (□) or LDI-1 C2 (▴) on budding of RSV Gag in 293 cells. The data are presented as the ratio of Pr76Gag in the media/lysate as a function of varying concentration of cotransfected expression plasmid. The LDI-1 WW and C2 data are derived from A. The extent of Gag budding in the absence of competing coexpressed proteins was set to 1.0. (C) Western blot analysis of expression of LDI-1 C2 and WW domains. Human 293/E cells were untransfected (lane 1) or transfected with 100 ng of AGP5 and 5 μg of LDI-1 C2 (lane 2) or LDI-1 WW (lane 3). Proteins were identified by Western blotting with a rabbit anti-HA antibody.
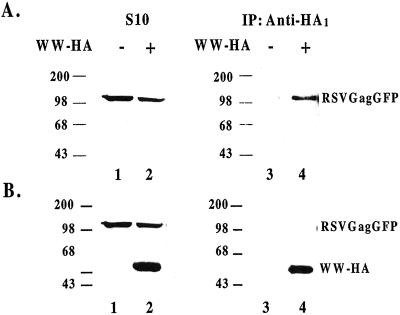
Coimmune Precipitation of Gag and LDI-1 from Cells.
To determine whether the LDI-1 WW domains inhibited Gag budding by competing with endogenous Nedd4-related protein(s) for binding to Gag, we attempted to coimmune precipitate the LDI-1 fragment with the four WW domains and Gag from cells. As shown in Fig. 5A, RSV Gag-EGFP was recognized by the anti-RSV p19Gag monoclonal antibody in the S10 fractions of cells untransfected and transfected with LDI-1 WW domains (lanes 1 and 2). Immune precipitation with rabbit anti-HA polyclonal antibody coprecipitated Gag only from those extracts expressing HA-tagged LDI-1 WW (compare lanes 3 and 4). Reprobing these blots with a different anti-HA antibody confirmed the expression of the LDI-1 WW domains containing fragment (Fig. 5B, lane 2) and its presence in the immune precipitate (lane 4). These results suggest that the LDI-1 fragment with the four WW domains inhibited RSV Gag release by interfering with formation of functionally competent budding complexes. We have confirmed these results by demonstrating that RSV Gag-EGFP and Nedd4 colocalized inside cells by using microscopy and fluorescent staining techniques (data not shown).
Figure 5.
Coimmune precipitation of RSV Gag and LDI-1 WW domains from cells. (A) Untransfected COS-1 cells (lanes 1 and 3) and cells transfected with pCMV-RSVGag-EGFP and HA-tagged LDI-1 WW domains (lanes 2 and 4) were lysed and proteins resolved and detected as described in legend to Fig. 2. Proteins in lanes 3 and 4 were immunoprecipitated with anti-HA rabbit sera before resolving the proteins by SDS/PAGE. (B) The nitrocellulose membranes described in A were reprobed in a Western analysis without stripping with a mouse anti-HA sera.
Discussion
LDI-1 shares the highest protein sequence homology with Xenopus Nedd4 (27) and the human KIAA0439 gene product (GenBank accession number AB007899), a ubiquitin protein ligase, with 85% and 88% sequence identity, respectively. This sequence homology suggests that LDI-1 is most likely the chicken homolog of these Nedd4-like proteins. Nedd4 was originally isolated by subtraction hybridization of mouse neuronal precursor cell cDNA, as a gene that showed developmentally regulated expression in the mouse embryonic central nervous system (28). Nedd4 is a protein of ≈120 kDa (29) and is found in mammals as well as Xenopus and yeast. It is a ubiquitin protein ligase composed of a putative membrane-binding C2 domain, three or four WW domains, an E2 protein binding region, and a C terminus ubiquitin protein ligase HECT domain (30). LDI-2 exhibited highest homology to human WWP1 (31), a ubiquitin protein ligase, and the mouse ubiquitin protein ligase, Itch, a gene whose knockout results in an aberrant inflammatory response that results in the death of young pups at about 4 months postnatal (32). LDI-1 and LDI-2 share only a 17% sequence identity. The coding regions of either chicken cDNA clones contain 4 WW domains each; however, only LDI-1 contains a discernable C2 domain. When compared with known E3 ubiquitin protein ligases, LDI-1 lacked the C-terminal half of the protein, which would contain the HECT domain, indicating that both clones were incomplete. Interestingly, a mouse homolog of the human KIAA0439 protein was recently shown, like Nedd4, to be potentially involved in the regulation of the epithelial sodium channel, possibly in a tissue-specific and/or signal-specific manner (33), which suggested localization at the plasma membrane.
Because LDI-1, which appears to play a role in RSV budding in this study, is a putative E3 ubiquitin protein ligase, ubiquitination of Gag may play a role in viral assembly and/or budding. Ubiquitination involves covalent ligation of ubiquitin to proteins via an isopeptide bond linking the C-terminal Gly of ubiquitin to the ɛ-amino group of Lys in the target protein. This process requires the sequential action of two or more enzymes. A ubiquitin-activating enzyme, E1, forms a high-energy thioester bond between a catalytic site Cys and ubiquitin in an ATP-dependent reaction. Ubiquitin is then transferred to a Lys residue of a target protein by the action of a ubiquitin-conjugating enzyme, E2, which in some cases works together with a ubiquitin protein ligase, E3 (34). Recent work has suggested that, whereas polyubiquitination marks proteins for proteasomal degradation, monoubiquitination targets proteins to a different pathway (35). A growing number of studies strongly suggest a role for ubiquitination of Gag in budding (see ref. 36 for a review). In 1990, Putterman et al. (37) reported the packaging of ubiquitin by avian leukosis virus and suggested the Gag polyprotein as the most likely viral protein candidate for ubiquitination, although direct ubiquitination of Gag was not demonstrated. More recently, Ott et al. (38) reported detecting small amounts of monoubiquitin-conjugated Gag products in HIV-1, SIV, and Moloney murine leukemia virus (Mo-MuLV) particles (p6Gag for HIV and SIV, and p12Gag for Mo-MuLV), as well as free ubiquitin in these viral particles. Strack et al. (39) demonstrated that diverse L domain sequences (including the RSV L domain) and particularly the L domain-like sequence from Ebola virus, promoted the ubiquitination of HIV-1 Gag. This study also reported that the overexpression of a human Nedd4 or a human Nedd4 in which the HECT domain active site Cys was mutated had no effect on retroviral budding. However, there are multiple members of the Nedd4 protein family in cells, and it is possible that the protein used by this group does not normally participate in HIV-1 Gag budding.
In separate studies, Schubert et al. (40) showed that proteasome inhibitors block HIV-1 Gag processing by the viral protease, via a mechanism other than direct inhibition of protease catalytic function. Patnaik et al. (41) showed that proteasome inhibitors also blocked RSV budding, resulting in the accumulation of arrays of fused particles on the cell surface, as detected by electron microscopy. This effect was reversed by the overexpression of ubiquitin and also by the fusion of ubiquitin to the C terminus of Gag. However, there was still a requirement of a functional L domain; ubiquitinated Gag, without an L domain, did not bud from cells (41). These reports are consistent both with a role for ubiquitination in budding and with our identification of a family of ubiquitinating enzymes, the E3 ubiquitin protein ligases, as cellular factors that interact with the RSV L domain required for budding. In a separate study, VerPlank et al. (26) used a yeast two-hybrid screen of a B cell library with HIV-1 Gag as bait and isolated the protein Tsg101, which interacts with the PTAPP L domain sequence of HIV-1 p6. The tsg101 gene was originally identified as a tumor suppressor (42). Tsg101 belongs to a group of inactive homologs of the ubiquitin-conjugating enzymes, E2, which may be dominant-negative regulators of E2 activity in cell-cycle control (43, 25). It also interacts with E3 ligases (44) and is involved in trafficking of plasma membrane proteins. Thus, the finding that two unique retroviral L domain sequences bind to proteins involved in ubiquitination suggests that the retroviral budding mechanism probably utilizes different components of the same cellular pathway.
Supplementary Material
Acknowledgments
We wish to thank Drs. Terry Copeland and Alex Wlodawer for the gift of the biotinylated peptides and Elena Brin for help with figures. This work was supported in part by National Institutes of Health Research Grants CA52047 (J.L.), GM48294 (C.C.), GM58271 (C.C.), and CA82177 (A.A.), awards from the Lymphoma and Leukemia Foundation (A.A.) and from the Leukemia Research Foundation (A.A.), and a United Nations Children Fund-Parke Davis Postdoctoral Fellowship (A.K.).
Abbreviations
- L domain
late assembly domain
- EIAV
equine infectious anemia virus
- LDI
late domain-interacting protein
- HA
hemagglutinin
- EBV
Epstein–Barr virus
- Nedd4
neuronal precursor cell-expressed developmentally down-regulated 4
- EGFP
enhanced green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bryant M, Ratner L. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wills J W, Craven R C. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Rein A, McClure M R, Rice N R, Luftig R B, Schultz A M. Proc Natl Acad Sci USA. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee S S, Hunter E. J Virol. 1987;61:1045–1053. doi: 10.1128/jvi.61.4.1045-1053.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weldon R A, Jr, Wills J W. J Virol. 1993;6:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang M, Orenstein J M, Martin M A, Freed E O. J Virol. 1995;6:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. J Virol. 1995;6:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puffer B A, Parent L J, Wills J W, Montelaro R C. J Virol. 1997;7:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. J Virol. 1994;6:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang Y, Cameron C E, Wills J W, Leis J. Virology. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harty R N, Paragas J, Sudol M, Palese P. J Virol. 1999;73:2921–2929. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jayakar H R, Murti K G, Whitt M A. J Virol. 2000;74:9818–9827. doi: 10.1128/jvi.74.21.9818-9827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bork P, Sudol M. Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 14.Chen H I, Sudol M. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Einbond A, Sudol M. FEBS Lett. 1996;38:1–8. doi: 10.1016/0014-5793(96)00263-3. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann K, Bucher P. FEBS Lett. 1995;35:153–157. doi: 10.1016/0014-5793(94)01415-w. [DOI] [PubMed] [Google Scholar]
- 17.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. J Virol. 1995;6:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puffer B A, Watkins S C, Montelaro R C. J Virol. 1998;72:10218–10221. doi: 10.1128/jvi.72.12.10218-10221.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staub O, Yeger H, Plant P J, Kim H, Ernst S A, Rotin D. Am J Physiol. 1997;272:1871–1880. doi: 10.1152/ajpcell.1997.272.6.C1871. [DOI] [PubMed] [Google Scholar]
- 21.Harvey K F, Dinudom A, Komwatana P, Jolliffe C N, Day M L, Parasivam G, Cook D I, Kumar S. J Biol Chem. 1999;27:12525–12530. doi: 10.1074/jbc.274.18.12525. [DOI] [PubMed] [Google Scholar]
- 22.Aiyar A, Tyree C, Sugden B. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aiyar A, Sugden B. J Biol Chem. 1998;273:33073–33081. doi: 10.1074/jbc.273.49.33073. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 26.VerPlank L, Bouamr F, LaGrassa T J, Agresta B, Kikonyogo A, Leis J, Carter C A. Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. . (First Published June 26, 2001; 10.1073/pnas131059198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rebhun J F, Pratt J H. DNA Seq. 1998;9:295–306. doi: 10.3109/10425179809008468. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Tomooka Y, Noda M. Biochem Biophys Res Comm. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 29.Anan T, Nagata Y, Koga H, Honda Y, Yabuki N, Miyamoto C, Kuwano A, Matsuda I, Endo F, Saya H, Nakao M. Genes Cells. 1998;3:751–763. doi: 10.1046/j.1365-2443.1998.00227.x. [DOI] [PubMed] [Google Scholar]
- 30.Staub O, Dho S, Henry P C, Correa J, Ishikawa T, McGlade J, Rotin D. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 31.Pirozzi G, McConnell S J, Uveges A J, Carter J M, Sparks A B, Kay B K, Fowlkes D M. J Biol Chem. 1997;272:14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- 32.Perry W L, Hustad C M, Swing D A, O'Sullivan T N, Jenkins N A, Copeland N G. Nat Genet. 1998;18:43–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- 33.Harvey K F, Dinudom A, Cook D I, Kumar S. J Biol Chem. 2001;276:8597–8601. doi: 10.1074/jbc.C000906200. [DOI] [PubMed] [Google Scholar]
- 34.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 35.Terrell J, Shih S, Dunn R, Hicke L. Mol Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- 36.Vogt V M. Proc Natl Acad Sci USA. 2000;97:12945–12947. doi: 10.1073/pnas.97.24.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Putterman D, Pepinsky R B, Vogt V M. Virology. 1990;176:633–637. doi: 10.1016/0042-6822(90)90035-p. [DOI] [PubMed] [Google Scholar]
- 38.Ott D E, Coren L V, Copeland T D, Kane B P, Johnson D G, Sowder R C, II, Yoshinaka Y, Oroszlan S, Arthur L, Henderson L. J Virol. 1998;72:2962–2968. doi: 10.1128/jvi.72.4.2962-2968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strack B, Calistri A, Accola M A, Palu G, Gottlinger H G. Proc Natl Acad Sci USA. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schubert U, Ott D E, Chertova E N, Welker R, Tessmer U, Princiotta M F, Bennick J R, Krausslich H-G, Yewdell J W. Proc Natl Acad Sci USA. 2000;97:13057–13062. doi: 10.1073/pnas.97.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patnaik A, Chau V, Wills J W. Proc Natl Acad Sci USA. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q, Chen C F, Chen Y, Chen P L, Lee W H. Cancer Res. 1997;57:4225–4228. [PubMed] [Google Scholar]
- 43.Koonin E V, Abagyan R A. Nat Genet. 1997;16:330–331. doi: 10.1038/ng0897-330. [DOI] [PubMed] [Google Scholar]
- 44.Li L, Liao J, Ruland J, Mak T W, Cohen S N. Proc Natl Acad Sci USA. 2000;98:1619–1624. doi: 10.1073/pnas.98.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.