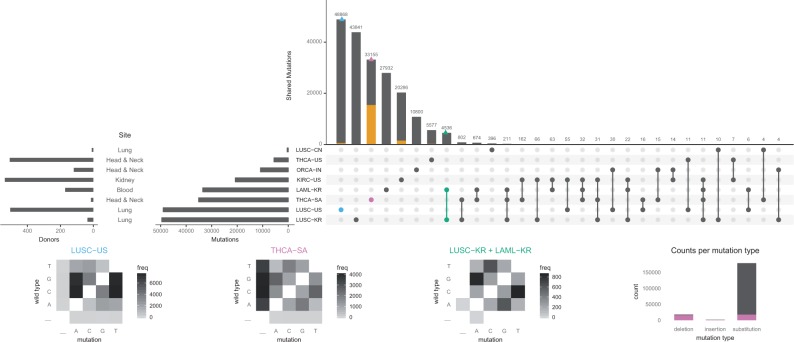
Fig. 1.
An UpSetR plot of variants across eight ICGC cancer studies with three intersection queries, one element query, four attribute plots, and two set metadata plots. Data for LUSC-KR and LAML-KR are based on whole-genome sequencing, all others on whole-exome sequencing. The three intersection queries are the one-way intersections of LUSC-US (blue) and THCA-SA (purple), and the two-way intersection of LUSC-KR and LAML-KR (green). The element query (yellow) selects mutations classified as deletions. Three custom transition/transversion plots display the relative frequency of substitution events for the intersection queries. The bar plot attribute plot displays the contribution of variants unique to the THCA-SA cohort (purple) to each mutation type. Set metadata is plotted to the left of the set size bar (charts)