Fig. 1.
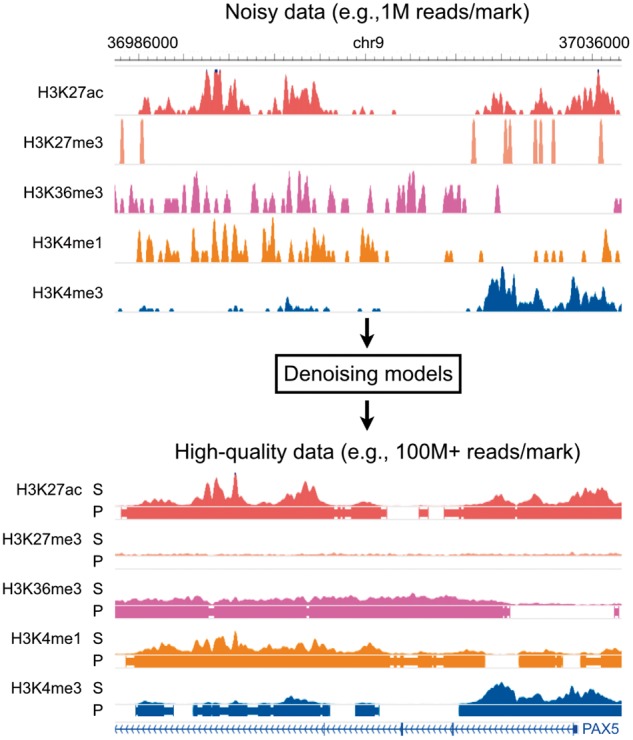
Overall model. Coda learns a transformation from noisy histone ChIP-seq data to a set of clean signal tracks and accurate peak calls. Top: a noisy signal track derived from 1M ChIP-seq reads per histone mark on the lymphoblastoid cell line GM12878. Bottom: a high-quality signal track derived from 100 + M ChIP-seq reads per histone mark from the same experiment. S, signal; P, peak calls
