Fig. 3.
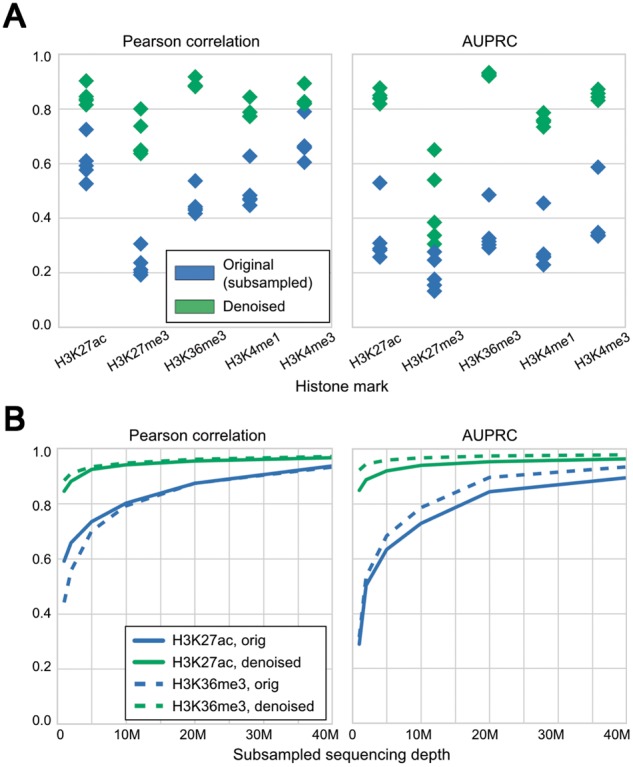
Coda removes noise from low-sequencing-depth experiments on lymphoblastoid cell lines derived from different individuals. (A) Compared with the signal from subsampled reads (blue), the denoised signal (green) shows greater correlation with the full signal (left) and more accurate peak-calling (right) across all cell lines. The model was trained on GM12878 and tested on different cell lines; within each column in the plot, each point is a single test cell line. (B) With 1M reads per mark, the denoised H3K27ac data are equivalent in quality to a dataset with 15M+ reads per mark, and the H3K36me3 data are equivalent in quality to a dataset with 25M+ reads per mark. Similar results hold for other marks. These results are from training on GM12878 and testing on GM18526
