Fig. 5.
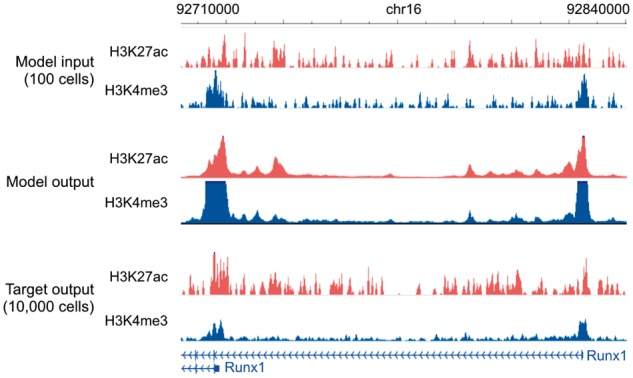
Genome browser tracks for low-cell-input experiments. We compare noisy signal obtained from 100 cells (top) with Coda’s output (middle) and the target, high-quality signal obtained from 10 000 cells (bottom) at the Runx1 gene in mouse hematopoietic stem and progenitor cells. The model was trained on MOWChIP-seq data generated from human LCL (GM12878) and captures two strong peaks at the promoters of the two isoform classes, removing much of the intervening noise. The signal tracks are in arcsinh units, with a scale of 0–40 for both histone marks
