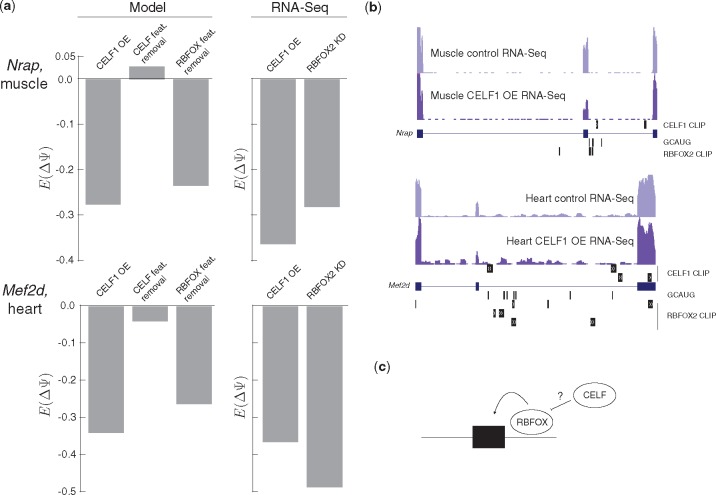
Fig. 5.
Splicing code models suggest indirect mechanism of key CELF1 regulatory targets. (a) Model predicted changes in exon inclusion for Nrap in muscle (top) and Mef2d in heart (bottom) upon CELF1 overexpression, removal of features related to the CELF family, or removal of features related to the RBFOX family (bars 1–3 from the left) as well as quantification of change in inclusion from RNA-Seq upon over expression of CELF1 or knockdown of RBFOX2 in myotubes (bars 4 and 5 from the left). (b) UCSC Genome Browser view of regulated cassette exons in Nrap (top) and Mef2d (bottom) showing locations of RNA-Seq reads in given conditions, CELF1 and RBFOX2 peaks, and the RBFOX family binding motif GCAUG. (c) Schematic representation of suggested regulatory relationship between RBFOX and CELF