Fig. 3.
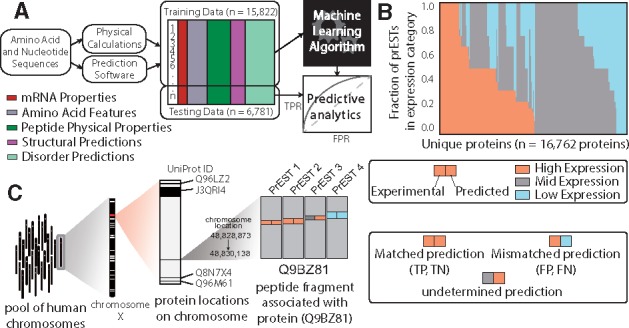
Machine learning-based approach to classify expression and solubility of protein fragments. (A) Classification workflow, starting with mRNA and amino acid sequences. The features described in Table 1 are generated for each PrEST and compiled into a feature matrix. Some of the data is used to train the models, and the rest is used to validate them. (B) The fraction of PrESTs in each expression level for each protein. (C) Multi-scale illustration of a protein in this study. Each protein is coded in a chromosome, and contains a number of PrESTs. Each PrEST has an experimental expression level and a predicted expression level from the machine learning algorithm
