Abstract
Background
There's no consensus about the difference between renal pelvic and ureteral tumors in terms of clinical features, pathological outcomes, epigenetic biomarkers and prognosis.
Methods
The data of 341 patients with renal pelvic tumors and 271 patients with ureteral tumors who underwent radical nephroureterectomy between 1999 and 2011 were retrospectively reviewed. The clinicopathologic features, gene promoters methylation status and oncologic outcomes were compared. Regression analysis was performed to identify oncologic prognosticators.
Results
Patients with ureteral tumors were relatively older (p = 0.002), and had higher likelihood of pre-operative renal insufficiency (p < 0.001), hypertension (p = 0.038) and hydronephrosis (P < 0.001), while in patients with renal pelvic tumors gross hematuria was more prevalent (p < 0.001). Renal pelvic tumors tended to exhibit non-organ-confined disease (p = 0.004) and larger tumor diameter (p = 0.001), while ureteral tumors had a higher likelihood of exhibiting high grade (p < 0.001) and sessile architecture (p = 0.023). Hypermethylated gene promoters were significantly more prevalent in renal pelvic tumors (p < 0.001), specifically for TMEFF2, GDF15, RASSF1A, SALL3 and ABCC6 (all p < 0.05). Tumor location failed to independently predict cancer-specific survival, overall survival, intravesical or contralateral recurrence (all p > 0.05), while gene methylation status was demonstrated to be an independent prognostic factor.
Conclusion
Renal pelvic tumors and ureteral tumors exhibited significant differences in clinicopathologic characteristics and epigenetic biomarkers. Gene promoter methylation might be an important mechanism in explaining distinct tumor patterns and behaviors in UTUC.
Keywords: Methylation, Prognosis, Radical nephroureterectomy (RNU), Renal pelvis, Upper tract urothelial carcinomas (UTUC), Ureter
Background
Urothelial carcinomas could be located anywhere throughout the whole urinary tract, e.g. renal pelvis, ureter, bladder and urethra [1]. Upper tract urothelial carcinoma refers to renal pelvic and ureteral tumors [2], with radical nephroureterectomy (RNU) and excision of the bladder cuff as the standard treatment [3].
Since both ureteral tumors and renal pelvic tumors originate from the urothelium, they have been traditionally classified as a single entity (UTUC) and managed in a relatively similar fashion, barring nephron-sparing approaches for more distally located tumors. In recent years there have been studies focusing on the impact of tumor location on prognosis [4–7], though evidence concerning clinical, pathological and genetic differences between renal pelvic and ureteral tumors remains scarce [8].
Microsatellite instability and hypermethylation have been proposed as key genetic differences between bladder cancer and UTUC [9–11], and we recently found gene promoter methylation status to hold biologic and prognostic significance in UTUC [12]. In the present study based on a large cohort of Chinese UTUC patients, we investigated the difference between renal pelvic and ureteral tumors in terms of clinical features, pathological outcomes, epigenetic biomarkers and prognosis.
Methods
Patient selection
Review board approve from Peking University First Hospital was acquired and all patients signed the informed consent to participate into the study. We evaluated consecutive Chinese UTUC patients who underwent RNU from 1999 to 2011 at Peking University First Hospital. We excluded patients with synchronous bilateral UTUC, distant metastasis prior to surgery or without complete follow-up data. Patients without available DNA from the surgical specimen for analysis of gene promoter methylation status were also excluded. Six hundred and-twelve patients were finally enrolled for analysis.
RNU including an extravesical excision of distal ureter by open Gibson incision was performed in all patients. No patients received neoadjuvant chemotherapy or prophylactic post-operative intravesical instillation (MMC or THP), while adjuvant chemotherapy for high-risk patients was administered at the treating physician’s discretion.
Patient evaluation
Computed tomography (CT) or magnetic resonance imaging (MRI), urological ultrasound, and cystoscopy were performed in all patients before surgery. Urinary cytology and ureteroscopy were used to help diagnosis. Renal function was assessed by estimated glomerular filtration rate (eGFR) calculated by Chinese population-specific equation: eGFR(ml/min/1.73m2) = 175 × Scr-1.234 × age-0.179 (× 0.79 if female) [13]. Ipsilateral hydronephrosis was determined pre-operatively.
Patients were categorized into 2 groups (renal pelvis versus ureter) in the current analysis based on the location of the main lesion on pathological specimen (e.g. the highest tumor stage). Pathological examination was performed according to standard procedures by a dedicated pathologist. Tumors were staged per the 2002 Union for International Cancer Control (UICC) TNM classification, and grading was evaluated per the World Health Organization (WHO) classification of 1973.
DNA extraction and methylation analysis
The procedure of DNA extraction and methylation analysis has been reported in a previous publication by our research group [12]. Based on the formalin-fixed paraffin-embedded tumor samples stored in our center, DNA samples were obtained and were treated for bisulfite transformation. Methylation-sensitive polymerase chain reaction (MSP) was used to analyze the gene promoters methylation status [14]. We used methylated human genomic DNA (Qiagen, Hilden, Germany) as positive control and water blanks with polymerase chain reaction mixtures as negative control. Based on previous literatures we did not detect the methylation status of the gene promoters in matched paracarcinoma tissues due to the limited methylation rates [15–20].
Follow-up schedule
Follow-up consisted of cystoscopy, chest X-ray, urine cytology, and serum creatinine every 3 months for the first 3 years and then once per year thereafter. Abdominal ultrasound or CT/MRI was performed to examine the contralateral upper urinary tract. Overall survival (OS), cancer specific survival (CSS), bladder recurrence and contralateral recurrence were documented and compared by tumor location. Bladder recurrence was defined as subsequent bladder tumor detected by cystoscopy and confirmed by pathologic examination, and contralateral recurrence was defined as urothelial carcinoma found in the contralateral upper urinary tract. Cause of death was determined by death certificates, by medical notes or by the patients’ responsible physicians.
Statistical analysis
Statistical analysis was carried by using R software i386 2.15.3 (R Foundation for Statistical Computing, http://www.r-project.org) and SPSS 20.0 (IBM Corp, Armonk, NY, USA). Categorical variables were tested by the Pearson’s test and Chi-square test, while variables with a continuous distribution were evaluated by the Mann-Whitney U test. Cox regression model was used for survival analysis, and Kaplan-Meier curves including log-rank test was employed. A single-sided p value of lower than 0.05 was regarded as statistical significance.
Results
Clinical characteristics
Overall, 612 patients with either renal pelvic tumor (n = 341; 55.7%) or ureteral tumor (n = 271; 44.3%) were included. The median age of the entire cohort of patients was 68 (interquartile range, IQR: 60–74) years, and 272 (44.4%) were female, with a male:female ratio of 1.25:1. Previous or concomitant bladder cancer was present in 67 patients (10.9%).
The clinical features are exhibited in Table 1, grouped by tumor location. Patients with ureteral tumors were relatively older (p = 0.002), and suffered from high likelihood of pre-operative renal insufficiency (p < 0.001), hypertension (p = 0.038) and hydronephrosis (P < 0.001), while in patients with renal pelvic tumors gross hematuria was more prevalent (p < 0.001).
Table 1.
Clinical and pathological characteristics of all UTUC patients stratified by tumor location
| Tumor location | Univariate analysis | ||||
|---|---|---|---|---|---|
| All | Renal pelvis | Ureter | Chi-square or Z | p value | |
| Patients, no. (%) | 612 (100) | 341 (55.7) | 271 (44.3) | ||
| Pre-operative characteristic | |||||
| Gender, no. (%) | 0.160 | 0.743 | |||
| Male | 340 (55.6) | 187 (54.8) | 153 (56.5) | ||
| Female | 272 (44.4) | 154 (45.2) | 118 (43.5) | ||
| Age, no. (%) | 4.929 | 0.027* | |||
| <70 | 340 (55.6) | 203 (59.5) | 137 (50.6) | ||
| ≥ 70 | 272 (44.4) | 138 (40.5) | 134 (49.4) | ||
| Age, mean ± SD | 65.29 ± 11.11 | 68.07 ± 10.20 | −3.173 | 0.002* | |
| Previous or concomitant bladder cancer, no. (%) | 1.931 | 0.193 | |||
| Absent | 545 (89.1) | 309 (90.6) | 236 (87.1) | ||
| Present | 67 (10.9) | 32 (9.4) | 35 (12.9) | ||
| Initial complaint, no. (%) | 24.205 | < 0.001* | |||
| Absent | 84 (13.7) | 26 (7.6) | 58 (21.4) | ||
| Present | 528 (86.3) | 315 (92.4) | 213 (78.6) | ||
| Gross hematuria, no. (%) | 65.132 | < 0.001* | |||
| Absent | 148 (24.2) | 40 (11.7) | 108 (39.9) | ||
| Present | 464 (75.8) | 301 (88.3) | 163 (60.1) | ||
| Preoperative renal function, no. (%) | 23.703 | < 0.001* | |||
| End-stage CKD (eGFR<15) | 34 (5.6) | 24 (7.0) | 10 (3.7) | ||
| Moderate CKD (60>eGFR≥15) | 198 (32.4) | 83 (24.3) | 115 (42.4) | ||
| Early CKD (eGFR≥60) | 378 (61.8) | 233 (68.3) | 145 (53.5) | ||
| eGFR, mean ± SD | 69.69 ± 30.11 | 62.43 ± 22.32 | −4.329 | < 0.001* | |
| Side, no. (%) | 1.115 | 0.329 | |||
| Left | 315 (51.5) | 182 (53.4) | 133 (49.1) | ||
| Right | 297 (48.5) | 159 (46.6) | 138 (50.9) | ||
| Hydronephrosis, no. (%) | 134.680 | < 0.001* | |||
| Absent | 273 (44.6) | 223 (65.4) | 50 (18.5) | ||
| Present | 339 (55.4) | 118 (34.6) | 221 (81.5) | ||
| Multifocality, no. (%) | 0.339 | 0.563 | |||
| Single | 472 (77.1) | 266 (78.0) | 206 (76.0) | ||
| Multiple | 140 (22.9) | 75 (22.0) | 65 (24.0) | ||
| Smoking, no. (%) | 0.050 | 0.836 | |||
| No | 497 (81.2) | 278 (81.5) | 219 (80.8) | ||
| Yes | 115 (18.8) | 63 (18.5) | 52 (19.2) | ||
| Alcohol, no. (%) | 0.697 | 0.452 | |||
| No | 539 (88.1) | 297 (87.1) | 242 (89.3) | ||
| Yes | 73 (11.9) | 44 (12.9) | 29 (10.7) | ||
| Diabetes, no. (%) | 0.249 | 0.661 | |||
| No | 511 (83.5) | 287 (84.2) | 224 (82.7) | ||
| Yes | 101 (16.5) | 54 (15.8) | 47 (17.3) | ||
| Hypertension, no. (%) | 4.454 | 0.038* | |||
| No | 363 (59.3) | 215 (63.0) | 148 (54.6) | ||
| Yes | 249 (40.7) | 126 (37.0) | 123 (45.4) | ||
| Pre-RNU ureteroscopy, no. (%) | 20.495 | < 0.001* | |||
| No | 536 (87.6) | 317 (93.0) | 219 (80.8) | ||
| Yes | 76 (12.4) | 24 (7.0) | 52 (19.2) | ||
| Pathological outcomes | |||||
| Architecture, no. (%) | 40.135 | < 0.001* | |||
| Papillary | 479 (78.3) | 299 (87.7) | 180 (66.4) | ||
| Sessile | 133 (21.7) | 42 (12.3) | 91 (33.6) | ||
| Tumor stage, no. (%) | 0.094 | 0.796 | |||
| Ta-T1 | 206 (33.7) | 113 (33.1) | 93 (34.3) | ||
| T2–4 | 406 (66.3) | 228 (66.9) | 178 (65.7) | ||
| Tumor grade, no. (%) | 31.628 | < 0.001* | |||
| G1 | 19 (3.1) | 4 (1.2) | 15 (5.5) | ||
| G2 | 334 (54.6) | 218 (63.9) | 116 (42.8) | ||
| G3 | 259 (42.3) | 119 (34.9) | 140 (51.7) | ||
| Lymph node status, no. (%) | 4.014 | 0.051 | |||
| N0 or Nx | 571 (93.3) | 312 (91.5) | 259 (95.6) | ||
| N+ | 41 (6.7) | 29 (8.5) | 12 (4.4) | ||
| Non-organ-confined disease, no. (%) | 8.257 | 0.004* | |||
| No | 412 (67.3) | 213 (62.5) | 199 (73.4) | ||
| Yes | 200 (32.7) | 128 (37.5) | 72 (26.6) | ||
| Tumor size, mean ± SD | 3.58 ± 2.15 | 3.27 ± 2.41 | −3.342 | 0.001* | |
| Histologic Subtype | |||||
| Tumor necrosis, no. (%) | 0.038 | 0.901 | |||
| No | 537 (87.7) | 300 (88.0) | 237 (87.5) | ||
| Yes | 75 (12.3) | 41 (12.0) | 34 (12.5) | ||
| Squamous metaplasia, no. (%) | 0.038 | 0.878 | |||
| No | 566 (92.5) | 316 (92.7) | 250 (92.3) | ||
| Yes | 46 (7.5) | 25 (7.3) | 21 (7.7) | ||
| Sarcomatoid metaplasia, no. (%) | 0.039 | 0.843 | |||
| No | 586 (95.8) | 327 (95.9) | 259 (95.6) | ||
| Yes | 26 (4.2) | 14 (4.1) | 12 (4.4) | ||
| Gland-like differentiation, no. (%) | 2.738 | 0.119 | |||
| No | 591 (96.6) | 333(97.7) | 258 (95.2) | ||
| Yes | 21 (3.4) | 8(2.3) | 13 (4.8) | ||
| Presence of CIS, no. (%) | 3.987 | 0.071 | |||
| No | 596 (97.4) | 336 (98.5) | 260 (95.9) | ||
| Yes | 16 (2.6) | 5 (1.5) | 11 (4.1) | ||
UTUC upper tract urothelial carcinoma, CKD chronic kidney disease, eGFR estimated glomerular filtration rate, RNU radical nephroureterectomy, CIS carcinoma in situ, SD standard deviation, HR Hazard Ratio, CI confidence interval
*Statistically significant
Pathological outcomes
The frequencies of muscle-invasive disease (≥pT2) and lymph node metastasis were comparable between groups; however, non-organ-confined tumors (≥pT3) were more prevalent in patients with renal pelvic tumors versus the ureteral tumor counterparts (p = 0.004). In concordance with this observation, sessile architecture and larger tumor size were more prevalent in patients with renal pelvic tumors as well (p < 0.001). G3 tumor grade, however, was present more often in ureteral tumors (p < 0.001). There were no differences in terms of squamous and glandular differentiation.
Molecular biomarkers
In 542 patients (88.6%) at least one methylated gene promoter was found, with a mean methylated genes number of 3.33 ± 2.31. Methylation was present significantly more frequently in renal pelvic tumors (Table 2), particularly with a higher rate of methylated TMEFF2, GDF15, RASSF1A, SALL3 and ABCC6 (all p < 0.05) (Fig. 1a). The mean number methylated genes in renal pelvic tumors was 3.71 ± 2.33, while in ureteral tumors was only 2.85 ± 2.19 (p < 0.001). Besides many patients with ureteral tumors presented with only very few methylated genes. (Fig. 1b).
Table 2.
Molecular biomarkers
| All | Renal pelvis | Ureter | Chi-square or Z | p value | |
|---|---|---|---|---|---|
| Patients, no. (%) | 612 (100) | 341 (55.7) | 271 (44.3) | ||
| TMEFF2, no. (%) | 6.717 | 0.011* | |||
| Unmethylated | 346 (56.5) | 177 (51.9) | 169 (62.4) | ||
| Methylated | 266 (43.5) | 164 (48.1) | 102 (37.6) | ||
| HSPA2, no. (%) | 3.172 | 0.083 | |||
| Unmethylated | 355 (58.0) | 187 (54.8) | 168 (62.0) | ||
| Methylated | 257 (42.0) | 154 (45.2) | 103 (38.0) | ||
| GDF15, no. (%) | 57.000 | < 0.001* | |||
| Unmethylated | 304 (49.7) | 123 (36.1) | 181 (66.8) | ||
| Methylated | 308 (50.3) | 218 (63.9) | 90 (33.2) | ||
| RASSF1A, no. (%) | 20.465 | < 0.001* | |||
| Unmethylated | 448 (73.2) | 225 (66.0) | 223 (82.3) | ||
| Methylated | 164 (26.8) | 116 (34.0) | 48 (17.7) | ||
| SALL3, no. (%) | 7.119 | 0.008* | |||
| Unmethylated | 403 (65.8) | 209 (61.3) | 194 (71.6) | ||
| Methylated | 209 (34.2) | 132 (38.7) | 77 (28.4) | ||
| VIM, no. (%) | 2.347 | 0.128 | |||
| Unmethylated | 219 (35.8) | 113 (33.1) | 106 (39.1) | ||
| Methylated | 393 (64.2) | 228 (66.9) | 165 (60.9) | ||
| ABCC6, no. (%) | 4.719 | 0.037* | |||
| Unmethylated | 523 (85.5) | 282 (82.7) | 241 (88.9) | ||
| Methylated | 89 (14.5) | 59 (17.3) | 30 (11.1) | ||
| CDH1, no. (%) | 0.208 | 0.728 | |||
| Unmethylated | 524 (85.6) | 290 (85.0) | 234 (86.3) | ||
| Methylated | 88 (14.4) | 51 (15.0) | 37 (13.7) | ||
| THBS1, no. (%) | 0.005 | 1.000 | |||
| Unmethylated | 457 (74.7) | 255 (74.8) | 202 (74.5) | ||
| Methylated | 155 (25.3) | 86 (25.2) | 69 (25.5) | ||
| BRCA1, no. (%) | 0.460 | 0.523 | |||
| Unmethylated | 504 (82.4) | 284 (83.3) | 220 (81.2) | ||
| Methylated | 108 (17.6) | 57 (16.7) | 51 (18.8) | ||
| Presence of hypermethylation in any gene, no. (%) | 9.420 | 0.003* | |||
| Unmethylated | 70 (11.4) | 27 (7.9) | 43 (15.9) | ||
| Methylated | 542 (88.6) | 314 (92.1) | 228 (84.1) | ||
| Mean methylated genes | 3.71 ± 2.33 | 2.85 ± 2.19 | −4.503 | < 0.001* | |
| Number of methylated genes, no. (%) | 17.202 | < 0.001* | |||
| 0–2 | 254 (41.5) | 118 (34.6) | 136 (50.2) | ||
| 3–5 | 243 (39.7) | 145 (42.5) | 98 (36.2) | ||
| 6–10 | 115 (18.8) | 78 (22.9) | 37 (13.7) | ||
| Number of methylated genes, no. (%) in Ta-1 | 11.251 | 0.004* | |||
| All | 206 (100) | 113 (54.9) | 93 (45.1) | ||
| 0–2 | 95 (46.1) | 41 (36.3) | 54 (58.1) | ||
| 3–5 | 80 (38.8) | 49 (43.4) | 31 (33.3) | ||
| 6–10 | 31 (15.0) | 23 (20.4) | 8 (8.6) | ||
| Number of methylated genes, no. (%) in T2–4 | 7.318 | 0.026* | |||
| All | 406 (100) | 228 (56.2) | 178 (43.8) | ||
| 0–2 | 159 (39.2) | 77 (33.8) | 82 (46.1) | ||
| 3–5 | 163 (40.1) | 96 (42.1) | 67 (37.6) | ||
| 6–10 | 84 (20.7) | 55 (24.1) | 29 (16.3) | ||
| Number of methylated genes, no. (%) in G1–2 | 18.433 | < 0.001* | |||
| All | 353 (100) | 222 (62.9) | 131 (37.1) | ||
| 0–2 | 156 (44.2) | 80 (36.0) | 76 (58.0) | ||
| 3–5 | 141 (39.9) | 97 (43.7) | 44 (33.6) | ||
| 6–10 | 56 (15.9) | 45 (20.3) | 11 (8.4) | ||
| Number of methylated genes, no. (%) in G3 | 4.449 | 0.108 | |||
| All | 259 (100) | 119 (45.9) | 140 (54.1) | ||
| 0–2 | 98 (37.8) | 38 (31.9) | 60 (42.9) | ||
| 3–5 | 102 (39.4) | 48 (40.3) | 54 (38.6) | ||
| 6–10 | 59 (22.8) | 33 (27.7) | 26 (18.6) |
*Statistically significant
UTUC upper tract urothelial carcinoma
Fig. 1.
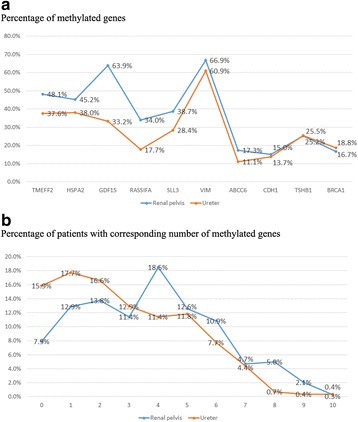
Distribution of aberrant methylated genes in renal pelvic and ureteral tumors (a) and the cumulative numbers of aberrant methylated genes (b)
In subgroup analysis based on tumor stage, renal pelvic tumors exhibited more methylated genes both in non-muscle-invasive and muscle-invasive diseases, while in subgroup analysis based on tumor grade, the difference was significant only in lower tumor stages (G1–2).
Oncologic outcomes
The median follow-up duration was 64 months. In all 210 (34.3%) patients died and 187 (30.6%) died secondary to urothelial cancer. The cumulative 5-year OS and CSS rates were 69.1% and 71.4%, respectively. Bladder recurrence was found in 174 (28.4%) patients, and 32 (5.2%) patients experienced contralateral recurrence.
By univariate analysis, there’s no relationship between tumor location (renal pelvis versus ureter) OS (p = 0.104), CSS (p = 0.071), bladder recurrence (p = 0.294) or contralateral recurrence (p = 0.871). (Fig. 2).
Fig. 2.
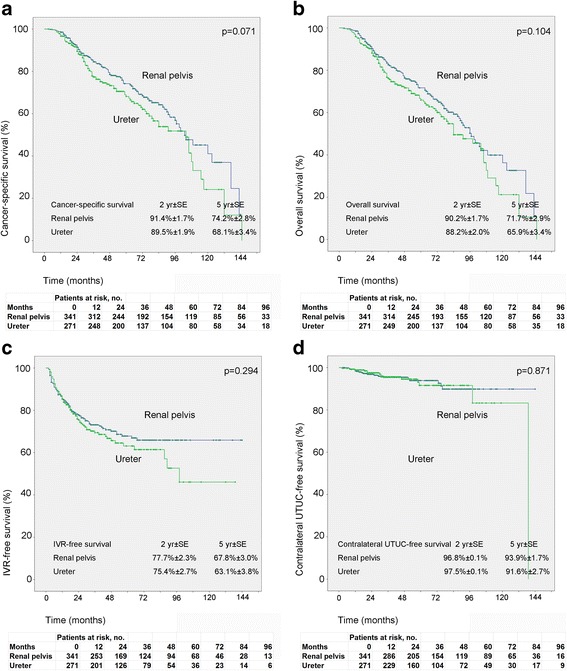
Estimated Kaplan-Meier overall survival (a) (p = 0.104), cancer specific survival (b) (p = 0.071), bladder recurrence-free survival (c) (p = 0.294) and contralateral carcinoma-free survival (d) (p = 0.871) curves stratified by tumor location
Other factors, including tumor stage, presence of hydronephrosis, and the methylation status of several genes were proved to be important predictive factors for survival. (Table 3). On Kaplan-Meier analysis, less cumulative number of methylated genes was correlated with better CSS, with mean CSS time of 101 months, 79 months and 77 months for patients with 0–2, 3–5 and 6–10 methylated genes, respectively (Fig. 3a). Though not statistically significant, a trend to higher risk for bladder recurrence in patients with less number of methylated genes (p = 0.081, Fig. 3b) was found. Besides the number of methylated genes (as continuous) was found to affect CSS (HR = 1.348, p = 0.003) and bladder recurrence (HR = 0.787, p = 0.026) in univariate analysis (Table 3 and 4).
Table 3.
Prognostic factors for cancer-specific survival in the entire cohort of UTUC patients and stratified by tumor location
| Variables | All patients (n = 612) | Renal pelvis (n = 341) | Ureter (n = 271) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UVA | MVA | UVA | MVA | UVA | MVA | |||||||||||||
| HR | 95%CI | p value | HR | 95%CI | p value | HR | 95%CI | p value | HR | 95%CI | p value | HR | 95%CI | p value | HR | 95%CI | p value | |
| Location (ureter vs renal pelvis) | 1.302 | 0.976–1.738 | 0.073 | |||||||||||||||
| Gender (men vs women) | 1.611 | 1.206–2.152 | 0.001* | 1.45 | 1.07–1.96 | 0.016* | 1.680 | 1.117–2.527 | 0.013* | 1.46 | 0.94–2.27 | 0.091 | 1.574 | 1.039–2.383 | 0.032* | 1.43 | 0.93–2.21 | 0.103 |
| Age (continuous) | 1.020 | 1.005–1.035 | 0.009* | 1.32 | 1.07–1.64 | 0.010* | 1.008 | 0.988–1.028 | 0.422 | 1.032 | 1.008–1.056 | 0.008* | 1.09 | 0.78–1.52 | 0.628 | |||
| Preoperative hydronephosis | 1.595 | 1.183–2.151 | 0.002* | 1.77 | 1.28–2.45 | 0.001* | 1.766 | 1.168–2.671 | 0.007* | 1.89 | 1.20–2.98 | 0.006* | 1.191 | 0.691–2.053 | 0.529 | |||
| Multifocality (presence of multiple foci vs absence) | 1.414 | 1.028–1.943 | 0.033* | 1.57 | 1.10–2.24 | 0.014* | 1.675 | 1.088–2.578 | 0.019* | 1.70 | 1.03–2.82 | 0.040* | 1.145 | 0.711–1.845 | 0.577 | |||
| Preoperative renal function(eGFR, continuous) | 0.996 | 0.991–1.001 | 0.119 | 1.000 | 0.993–1.007 | 0.993 | 0.989 | 0.979–0.998 | 0.021* | 0.77 | 0.55–1.06 | 0.106 | ||||||
| Previous or concomitant BT (presence vs absence) | 1.544 | 1.019–2.339 | 0.041* | 1.53 | 0.97–2.41 | 0.070 | 1.980 | 1.099–3.568 | 0.023* | 2.03 | 1.04–3.94 | 0.037* | 1.183 | 0.657–2.132 | 0.575 | |||
| Gross hematuria (presence vs absence) | 0.913 | 0.650–1.282 | 0.599 | 0.625 | 0.353–1.104 | 0.105 | 1.329 | 0.845–2.090 | 0.218 | |||||||||
| Smoke (presence vs absence) | 1.102 | 0.765–1.588 | 0.601 | 1.242 | 0.757–2.037 | 0.391 | 0.985 | 0.572–1.698 | 0.958 | |||||||||
| Alcohol (presence vs absence) | 1.108 | 0.726–1.690 | 0.634 | 1.546 | 0.912–2.619 | 0.105 | 0.713 | 0.344–1.477 | 0.363 | |||||||||
| Diabetes (presence vs absence) | 0.891 | 0.592–1.340 | 0.579 | 0.994 | 0.563–1.756 | 0.984 | 0.797 | 0.442–1.437 | 0.451 | |||||||||
| Hypertension (presence vs absence) | 1.180 | 0.879–1.584 | 0.270 | 1.056 | 0.689–1.618 | 0.802 | 1.230 | 0.810–1.868 | 0.332 | |||||||||
| Preoperative ureteroscopy | 0.616 | 0.383–0.992 | 0.046* | 0.72 | 0.44–1.18 | 0.194 | 0.234 | 0.058–0.951 | 0.042* | 0.24 | 0.06–1.03 | 0.055 | 0.663 | 0.387–1.137 | 0.135 | |||
| Tumor stage^ (T4 vs T3 vs T2 vs T1 vs Ta) | 1.725 | 1.443–2.061 | < 0.001* | 2.42 | 1.56–3.76 | < 0.001* | 1.514 | 1.197–1.915 | 0.001* | 1.83 | 1.04–3.21 | 0.035* | 2.288 | 1.716–3.050 | < 0.001* | 2.53 | 1.46–4.38 | 0.001* |
| Tumor grade^ (G3 vs G2 vs G1) | 1.593 | 1.222–2.075 | 0.001* | 0.69 | 0.36–1.34 | 0.274 | 1.284 | 0.868–1.900 | 0.211 | 1.856 | 1.271–2.710 | 0.001* | 1.30 | 0.47–3.61 | 0.611 | |||
| Lymph node status (N+ vs Nx vs N-) | 2.524 | 1.583–4.023 | < 0.001* | 1.82 | 1.08–3.07 | 0.024* | 2.863 | 1.615–5.074 | < 0.001* | 2.49 | 1.26–4.92 | 0.009* | 2.356 | 1.024–5.417 | 0.044* | 1.18 | 0.49–2.84 | 0.713 |
| Architecture (presence of sessile vs absence) | 1.974 | 1.437–2.713 | < 0.001* | 1.38 | 0.92–2.07 | 0.125 | 2.105 | 1.242–3.566 | 0.006* | 1.20 | 0.63–2.28 | 0.584 | 1.811 | 1.186–2.766 | 0.006* | 0.98 | 0.55–1.75 | 0.951 |
| CIS (presence of sessile vs absence) | 1.027 | 0.480–2.202 | 0.994 | 1.386 | 0.424–4.535 | 0.590 | 0.808 | 0.296–2.210 | 0.678 | |||||||||
| Necrosis(presence vs absence) | 1.925 | 1.302–2.846 | 0.001* | 1.36 | 0.84–2.18 | 0.207 | 1.606 | 0.905–2.850 | 0.105 | 2.352 | 1.375–4.025 | 0.002* | 1.36 | 0.69–2.70 | 0.376 | |||
| Squamous metaplasia (presence vs absence) | 1.783 | 1.081–2.943 | 0.024* | 1.45 | 0.85–2.48 | 0.171 | 2.123 | 1.063–4.241 | 0.033* | 2.31 | 1.06–5.02 | 0.034* | 1.485 | 0.716–3.079 | 0.288 | |||
| Sarcomatoid metaplasia (presence vs absence) | 2.595 | 1.526–4.413 | < 0.001* | 0.79 | 0.40–1.56 | 0.493 | 2.541 | 1.171–5.513 | 0.018* | 1.03 | 0.40–2.62 | 0.955 | 2.629 | 1.266–5.459 | 0.010* | 1.00 | 0.41–2.45 | 0.993 |
| Gland-like differentiation (presence vs absence) | 1.963 | 0.965–3.995 | 0.063 | 3.394 | 1.239–9.296 | 0.017* | 2.08 | 0.68–6.34 | 0.197 | 1.229 | 0.449–3.362 | 0.688 | ||||||
| Tumor size (continuous) | 1.172 | 1.112–1.236 | < 0.001* | 1.17 | 1.04–1.32 | 0.010* | 1.163 | 1.070–1.264 | < 0.001* | 1.16 | 0.93–1.44 | 0.188 | 1.184 | 1.110–1.264 | < 0.001* | 1.26 | 1.06–1.49 | 0.008* |
| TMEFF2 (methylated vs unmethylated) | 1.812 | 1.353–2.427 | < 0.001* | 1.67 | 1.12–2.50 | 0.012* | 1.634 | 1.085–2.459 | 0.019* | 1.16 | 0.70–1.92 | 0.562 | 2.189 | 1.434–3.340 | < 0.001* | 1.84 | 0.97–3.50 | 0.061 |
| HSPA2 (methylated vs unmethylated) | 1.815 | 1.349–2.442 | < 0.001* | 1.52 | 1.03–2.24 | 0.036* | 2.064 | 1.365–3.119 | 0.001* | 1.40 | 0.86–2.28 | 0.180 | 1.698 | 1.097–2.626 | 0.017* | 1.08 | 0.60–1.97 | 0.793 |
| GDF15 (methylated vs unmethylated) | 1.242 | 0.930–1.660 | 0.142 | 1.575 | 1.025–2.421 | 0.038* | 1.24 | 0.73–2.12 | 0.426 | 1.152 | 0.741–1.793 | 0.530 | ||||||
| RASSF1A (methylated vs unmethylated) | 1.383 | 1.002–1.908 | 0.049* | 1.15 | 0.78–1.70 | 0.477 | 1.271 | 0.824–1.961 | 0.279 | 1.796 | 1.102–2.929 | 0.019* | 1.57 | 0.87–2.82 | 0.135 | |||
| SALL3 (methylated vs unmethylated) | 1.214 | 0.887–1.662 | 0.226 | 0.887 | 0.565–1.392 | 0.602 | 1.853 | 1.190–2.885 | 0.006* | 1.58 | 0.93–2.68 | 0.094 | ||||||
| VIM (methylated vs unmethylated) | 1.360 | 1.002–1.847 | 0.049* | 0.99 | 0.68–1.44 | 0.941 | 1.630 | 1.041–2.550 | 0.033* | 1.37 | 0.81–2.32 | 0.243 | 1.208 | 0.786–1.857 | 0.388 | |||
| ABCC6 (methylated vs unmethylated) | 1.430 | 0.928–2.203 | 0.105 | 1.206 | 0.682–2.134 | 0.519 | 2.283 | 1.165–4.476 | 0.016* | 1.51 | 0.67–3.38 | 0.317 | ||||||
| CDH1 (methylated vs unmethylated) | 1.178 | 0.766–1.812 | 0.456 | 1.112 | 0.618–2.001 | 0.724 | 1.401 | 0.741–2.649 | 0.300 | |||||||||
| THBS1 (methylated vs unmethylated) | 1.131 | 0.811–1.577 | 0.468 | 0.877 | 0.534–1.439 | 0.603 | 1.415 | 0.899–2.227 | 0.133 | |||||||||
| BRCA1 (methylated vs unmethylated) | 0.851 | 0.565–1.280 | 0.438 | 0.678 | 0.361–1.272 | 0.226 | 1.026 | 0.596–1.765 | 0.927 | |||||||||
| No. methylated genes (continuous) | 1.348 | 1.107–1.641 | 0.003* | 0.62 | 0.30–1.28 | 0.193 | 1.225 | 0.930–1.613 | 0.149 | 1.646 | 1.234–2.196 | 0.001* | 0.58 | 0.18–1.82 | 0.351 | |||
UVA univariate analysis, MVA multivariate analysis, eGFR estimated glomerular filtration rate, UTUC upper tract urothelial carcinoma, BT bladder tumor, CIS carcinoma in situ, HR Hazard Ratio, CI confidence interval
*Statistically significant
Fig. 3.
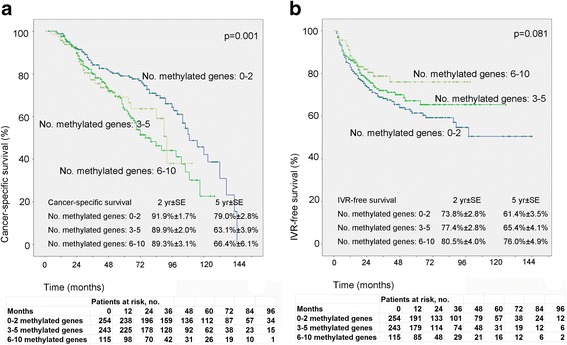
Estimated Kaplan-Meier cancer specific survival (a) (p = 0.001) and bladder recurrence-free survival (b) (p = 0.081) curves stratified by numbers of methylated genes (0–2 versus 3–5 versus 6–10)
Table 4.
Prognostic factors for bladder recurrence in the entire cohort of UTUC patients and stratified by tumor location
| Variables | All patients (n = 612) | Renal pelvis (n = 341) | Ureter (n = 271) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UVA | MVA | UVA | MVA | UVA | MVA | |||||||||||||
| HR | 95%CI | p value | HR | 95%CI | p value | HR | 95%CI | p value | HR | 95%CI | p value | HR | 95%CI | p value | HR | 95%CI | p value | |
| Location (ureter vs renal pelvis) | 1.172 | 0.870–1.579 | 0.297 | |||||||||||||||
| Gender (men vs women) | 1.150 | 0.854–1.550 | 0.357 | 1.279 | 0.850–1.924 | 0.239 | 1.035 | 0.668–1.606 | 0.876 | |||||||||
| Age (continuous) | 0.987 | 0.974–1.000 | 0.050 | 0.983 | 0.966–1.000 | 0.055 | 0.989 | 0.970–1.010 | 0.989 | |||||||||
| Preoperative hydronephosis | 1.173 | 0.868–1.585 | 0.298 | 1.141 | 0.745–1.748 | 0.545 | 1.050 | 0.608–1.815 | 0.861 | |||||||||
| Multifocality (presence of multiple foci vs absence) | 1.732 | 1.260–2.381 | 0.001* | 1.42 | 1.01–2.01 | 0.045* | 1.833 | 1.185–2.835 | 0.006* | 1.75 | 1.13–2.72 | 0.012* | 1.610 | 1.011–2.566 | 0.045* | 1.44 | 0.86–2.43 | 0.167 |
| Preoperative renal function(eGFR, continuous) | 0.999 | 0.993–1.004 | 0.671 | 0.999 | 0.993–1.006 | 0.876 | 0.999 | 0.989–1.009 | 0.887 | |||||||||
| Previous or concomitant BT (presence vs absence) | 1.900 | 1.267–2.850 | 0.002* | 1.47 | 0.95–2.28 | 0.081 | 1.710 | 0.931–3.138 | 0.084 | 2.034 | 1.176–3.517 | 0.011* | 1.39 | 0.75–2.58 | 0.293 | |||
| Gross hematuria (presence vs absence) | 1.108 | 0.773–1.588 | 0.577 | 1.102 | 0.572–2.125 | 0.772 | 1.268 | 0.802–2.005 | 0.310 | |||||||||
| Smoke (presence vs absence) | 0.946 | 0.644–1.388 | 0.775 | 1.097 | 0.655–1.836 | 0.726 | 0.784 | 0.441–1.394 | 0.407 | |||||||||
| Alcohol (presence vs absence) | 0.771 | 0.473–1.257 | 0.297 | 0.901 | 0.480–1.692 | 0.746 | 0.622 | 0.286–1.351 | 0.230 | |||||||||
| Diabetes (presence vs absence) | 1.086 | 0.736–1.601 | 0.679 | 0.842 | 0.469–1.515 | 0.567 | 1.414 | 0.837–2.388 | 0.196 | |||||||||
| Hypertension (presence vs absence) | 0.774 | 0.566–1.059 | 0.109 | 0.916 | 0.595–1.412 | 0.692 | 0.614 | 0.391–0.967 | 0.035* | 0.71 | 0.44–1.15 | 0.163 | ||||||
| Preoperative ureteroscopy | 1.631 | 1.111–2.395 | 0.012* | 1.25 | 0.83–1.87 | 0.285 | 2.087 | 1.137–3.829 | 0.018* | 1.62 | 0.87–3.02 | 0.126 | 1.319 | 0.794–2.189 | 0.285 | |||
| Tumor stage^ (T4 vs T3 vs T2 vs T1 vs Ta) | 0.861 | 0.731–1.014 | 0.074 | 0.956 | 0.772–1.185 | 0.683 | 0.744 | 0.574–0.965 | 0.026* | 0.81 | 0.54–1.20 | 0.292 | ||||||
| Tumor grade^ (G3 vs G2 vs G1) | 0.655 | 0.504–0.851 | 0.002* | 0.54 | 0.31–0.93 | 0.027* | 0.515 | 0.336–0.789 | 0.002* | 0.30 | 0.13–0.71 | 0.006* | 0.777 | 0.555–1.089 | 0.143 | |||
| Lymph node status (N+ vs Nx vs N-) | 0.326 | 0.121–0.879 | 0.027* | 0.45 | 0.17–1.23 | 0.120 | 0.491 | 0.180–1.338 | 0.164 | 0.046 | 0.000–5.017 | 0.199 | ||||||
| Architecture (presence of sessile vs absence) | 0.718 | 0.479–1.075 | 0.108 | 0.721 | 0.349–1.490 | 0.377 | 0.644 | 0.389–1.067 | 0.087 | |||||||||
| CIS (presence of sessile vs absence) | 1.604 | 0.789–3.262 | 0.192 | 1.498 | 0.369–6.086 | 0.572 | 1.555 | 0.676–3.573 | 0.299 | |||||||||
| Necrosis(presence vs absence) | 1.164 | 0.750–1.806 | 0.498 | 1.532 | 0.881–2.664 | 0.131 | 0.810 | 0.390–1.682 | 0.572 | |||||||||
| Squamous metaplasia(presence vs absence) | 0.626 | 0.308–1.274 | 0.196 | 0.630 | 0.231–1.720 | 0.367 | 0.609 | 0.223–1.665 | 0.334 | |||||||||
| Sarcomatoid metaplasia (presence vs absence) | 0.490 | 0.182–1.321 | 0.159 | 0.465 | 0.114–1.887 | 0.284 | 0.541 | 0.133–2.204 | 0.392 | |||||||||
| Gland-like differentiation (presence vs absence) | 0.576 | 0.184–1.806 | 0.344 | 0.619 | 0.086–4.452 | 0.634 | 0.509 | 0.125–2.073 | 0.346 | |||||||||
| Tumor size (continuous) | 0.920 | 0.850–0.996 | 0.039* | 0.91 | 0.78–1.06 | 0.213 | 0.967 | 0.872–1.072 | 0.520 | 0.875 | 0.771–0.991 | 0.036* | 0.83 | 0.64–1.08 | 0.170 | |||
| TMEFF2 (methylated vs unmethylated) | 0.714 | 0.521–0.978 | 0.036* | 0.91 | 0.60–1.38 | 0.657 | 0.839 | 0.554–1.268 | 0.404 | 0.593 | 0.358–0.984 | 0.043* | 1.00 | 0.50–2.00 | 0.999 | |||
| HSPA2 (methylated vs unmethylated) | 0.704 | 0.511–0.968 | 0.031* | 0.82 | 0.55–1.24 | 0.348 | 0.792 | 0.519–1.207 | 0.278 | 0.626 | 0.380–1.033 | 0.067 | ||||||
| GDF15 (methylated vs unmethylated) | 0.823 | 0.611–1.110 | 0.203 | 0.936 | 0.616–1.421 | 0.755 | 0.738 | 0.456–1.196 | 0.217 | |||||||||
| RASSF1A (methylated vs unmethylated) | 0.598 | 0.407–0.878 | 0.009* | 0.69 | 0.45–1.07 | 0.095 | 0.786 | 0.499–1.237 | 0.298 | 0.318 | 0.138–0.731 | 0.007* | 0.41 | 0.17–0.97 | 0.042* | |||
| SALL3 (methylated vs unmethylated) | 0.725 | 0.519–1.013 | 0.059 | 0.669 | 0.427–1.048 | 0.079 | 0.837 | 0.505–1.386 | 0.489 | |||||||||
| VIM (methylated vs unmethylated) | 0.862 | 0.636–1.167 | 0.336 | 1.302 | 0.831–2.041 | 0.249 | 0.567 | 0.367–0.875 | 0.010* | 0.64 | 0.38–1.06 | 0.081 | ||||||
| ABCC6 (methylated vs unmethylated) | 0.805 | 0.499–1.297 | 0.373 | 1.007 | 0.578–1.752 | 0.981 | 0.508 | 0.185–1.392 | 0.188 | |||||||||
| CDH1 (methylated vs unmethylated) | 0.681 | 0.413–1.124 | 0.133 | 0.870 | 0.474–1.597 | 0.654 | 0.466 | 0.188–1.155 | 0.099 | |||||||||
| THBS1 (methylated vs unmethylated) | 0.960 | 0.678–1.359 | 0.818 | 1.044 | 0.651–1.675 | 0.858 | 0.872 | 0.522–1.457 | 0.601 | |||||||||
| BRCA1 (methylated vs unmethylated) | 0.977 | 0.659–1.448 | 0.908 | 0.858 | 0.485–1.515 | 0.597 | 1.131 | 0.655–1.955 | 0.659 | |||||||||
| No. methylated genes(continuous) | 0.787 | 0.637–0.972 | 0.026* | 1.14 | 0.56–2.34 | 0.718 | 0.906 | 0.688–1.193 | 0.483 | 0.656 | 0.460–0.935 | 0.020* | 0.96 | 0.34–2.72 | 0.934 | |||
*Statistically significant
UVA univariate analysis, MVA multivariate analysis, eGFR estimated glomerular filtration rate, UTUC upper tract urothelial carcinoma, BT bladder tumor, CIS carcinoma in situ, HR Hazard Ratio, CI confidence interval
Sub-group analysis demonstrated differences in oncologic prognosticators for CSS and bladder recurrence based on tumor location (Table 3 and 4). Rerunning the dataset by dividing patients into renal pelvic tumors only (n = 304), ureteral tumors only (n = 267) and both renal pelvic and ureteral tumors (n = 41) did not change the results (Table 5).
Table 5.
Comparison in patients with renal pelvis tumor only and with ureteral tumor only
| Location | Comparison between three groups | Comparison after excluding cases in both locations | ||||||
|---|---|---|---|---|---|---|---|---|
| All | Renal pelvis only | Ureter only | Both locations | Chi-square or Z | p value | Chi-square or Z | p value | |
| Patients, no. (%) | 612 (100) | 304 (49.7) | 267 (43.6) | 41 (6.7) | ||||
| Pre-operative characteristic | ||||||||
| Gender, no. (%) | 1.595 | 0.450 | 0.495 | 0.501 | ||||
| Male | 340 (55.6) | 163 (53.6) | 151 (56.6) | 26 (63.4) | ||||
| Female | 272 (44.4) | 141 (46.4) | 116 (43.4) | 15 (36.6) | ||||
| Age, no. (%) | 5.554 | 0.062 | 5.391 | 0.023* | ||||
| <70 | 340 (55.6) | 182 (59.9) | 134 (50.2) | 24 (58.5) | ||||
| ≥ 70 | 272 (44.4) | 122 (40.1) | 133 (49.8) | 17 (41.5) | ||||
| Age, mean ± SD | 65.09 ± 11.32 | 68.12 ± 10.22 | 66.52 ± 10.79 | 11.059 | 0.004* | −3.298 | 0.001* | |
| Previous or concomitant bladder cancer, no. (%) | 31.791 | < 0.001* | 8.721 | 0.004* | ||||
| Absent | 545 (89.1) | 286 (94.1) | 232 (86.9) | 27 (65.9) | ||||
| Present | 67 (10.9) | 18 (5.9) | 35 (13.1) | 14 (34.1) | ||||
| Initial complaint, no. (%) | 23.992 | < 0.001* | 23.745 | < 0.001* | ||||
| Absent | 84 (13.7) | 22 (7.2) | 57 (21.3) | 5 (12.2) | ||||
| Present | 528 (86.3) | 282 (92.8) | 210 (78.7) | 36 (87.8) | ||||
| Gross hematuria, no. (%) | 66.717 | < 0.001* | 65.579 | < 0.001* | ||||
| Absent | 148 (24.2) | 33 (10.9) | 107 (40.1) | 8 (19.5) | ||||
| Present | 464 (75.8) | 271 (89.1) | 160 (59.9) | 33 (80.5) | ||||
| Preoperative renal function, no. (%) | 39.081 | < 0.001* | 29.841 | < 0.001* | ||||
| End-stage CKD (eGFR<15) | 34 (5.6) | 21 (6.9) | 10 (3.7) | 3 (7.3) | ||||
| Moderate CKD (60>eGFR≥15) | 198 (32.4) | 64 (21.1) | 112 (41.9) | 22 (53.7) | ||||
| Early CKD (eGFR≥60) | 378 (61.8) | 218 (71.7) | 144 (53.9) | 16 (39.0) | ||||
| eGFR, mean ± SD | 71.30 ± 29.38 | 62.63 ± 22.32 | 55.80 ± 31.99 | 34.160 | < 0.001* | −5.108 | < 0.001* | |
| Hydronephrosis, no. (%) | 156.085 | < 0.001* | 151.247 | < 0.001* | ||||
| Absent | 273 (44.6) | 212 (69.7) | 49 (18.4) | 12 (29.3) | ||||
| Present | 339 (55.4) | 92 (30.3) | 218 (81.6) | 29 (70.7) | ||||
| Multifocality, no. (%) | 156.779 | < 0.001* | 10.618 | < 0.001* | ||||
| Single | 472 (77.1) | 266 (87.5) | 206 (77.2) | 0 | ||||
| Multiple | 140 (22.9) | 38 (12.5) | 61 (22.8) | 41 (100) | ||||
| Pathological outcomes | ||||||||
| Architecture, no. (%) | 39.792 | < 0.001* | 39.811 | < 0.001* | ||||
| Papillary | 479 (78.3) | 269 (88.5) | 178 (66.7) | 32 (78.0) | ||||
| Sessile | 133 (21.7) | 35 (12.5) | 89 (33.3) | 9 (22.0) | ||||
| Tumor stage, no. (%) | 0.160 | 0.923 | 0.155 | 0.723 | ||||
| Ta-T1 | 206 (33.7) | 100 (32.9) | 92 (34.5) | 14 (34.1) | ||||
| T2–4 | 406 (66.3) | 204 (67.1) | 175 (65.5) | 27 (65.9) | ||||
| Tumor grade, no. (%) | 30.572 | < 0.001* | 28.242 | < 0.001* | ||||
| G1 | 19 (3.1) | 4 (1.3) | 15 (5.6) | 0 | ||||
| G2 | 334 (54.6) | 214 (70.4) | 115 (43.1) | 25 (61.0) | ||||
| G3 | 259 (42.3) | 106 (34.9) | 137 (51.3) | 16 (39.0) | ||||
| Lymph node status, no. (%) | 3.772 | 0.152 | 3.769 | 0.064 | ||||
| N0 or Nx | 571 (93.3) | 278 (91.4) | 255 (95.5) | 38 (92.7) | ||||
| N+ | 41 (6.7) | 26 (8.6) | 12 (4.5) | 3 (7.3) | ||||
| Non-organ-confined disease, no. (%) | 10.339 | 0.006* | 9.592 | 0.002* | ||||
| No | 412 (67.3) | 186 (61.2) | 196 (73.4) | 30 (73.2) | ||||
| Yes | 200 (32.7) | 118 (38.8) | 71 (26.6) | 11 (26.8) | ||||
| Tumor size, mean ± SD | 3.56 ± 1.94 | 3.25 ± 2.40 | 3.89 ± 3.39 | 13.014 | 0.001* | −3.695 | < 0.001* | |
| Methylation status | ||||||||
| TMEFF2, no. (%) | 6.972 | 0.031* | 6.481 | 0.011* | ||||
| Unmethylated | 346 (56.5) | 158 (52.0) | 167 (62.5) | 21 (51.2) | ||||
| Methylated | 266 (43.5) | 146 (48.0) | 100 (37.5) | 20 (48.8) | ||||
| HSPA2, no. (%) | 3.398 | 0.183 | 3.064 | 0.089 | ||||
| Unmethylated | 355 (58.0) | 167 (54.9) | 166 (62.2) | 22 (53.7) | ||||
| Methylated | 257 (42.0) | 137 (45.1) | 101 (37.8) | 19 (46.3) | ||||
| GDF15, no. (%) | 56.507 | < 0.001* | 56.310 | < 0.001* | ||||
| Unmethylated | 304 (49.7) | 107 (35.2) | 178 (66.7) | 19 (46.3) | ||||
| Methylated | 308 (50.3) | 197 (64.8) | 89 (33.3) | 22 (53.7) | ||||
| RASSF1A, no. (%) | 22.562 | < 0.001* | 22.341 | < 0.001* | ||||
| Unmethylated | 448 (73.2) | 197 (64.8) | 220 (82.4) | 31 (75.6) | ||||
| Methylated | 164 (26.8) | 107 (35.2) | 47 (17.6) | 10 (24.4) | ||||
| SALL3, no. (%) | 9.797 | 0.007* | 6.982 | 0.010* | ||||
| Unmethylated | 403 (65.8) | 188 (61.8) | 193 (72.3) | 22 (53.7) | ||||
| Methylated | 209 (34.2) | 116 (38.2) | 74 (27.7) | 19 (46.3) | ||||
| VIM, no. (%) | 3.367 | 0.186 | 1.819 | 0.192 | ||||
| Unmethylated | 219 (35.8) | 103 (33.9) | 105 (39.3) | 11 (26.8) | ||||
| Methylated | 393 (64.2) | 201 (66.1) | 162 (60.7) | 30 (73.2) | ||||
| ABCC6, no. (%) | 6.282 | 0.043* | 6.119 | 0.016* | ||||
| Unmethylated | 523 (85.5) | 250 (82.2) | 239 (89.5) | 34 (82.9) | ||||
| Methylated | 89 (14.5) | 54 (17.8) | 28 (10.5) | 7 (17.1) | ||||
| CDH1, no. (%) | 1.054 | 0.590 | 0.116 | 0.809 | ||||
| Unmethylated | 524 (85.6) | 260 (85.5) | 231 (86.5) | 33 (80.5) | ||||
| Methylated | 88 (14.4) | 44 (14.5) | 36 (13.5) | 8 (19.5) | ||||
| THBS1, no. (%) | 1.041 | 0.594 | 0.096 | 0.772 | ||||
| Unmethylated | 457 (74.7) | 230 (75.7) | 199 (74.5) | 28 (68.3) | ||||
| Methylated | 155 (25.3) | 74 (24.3) | 68 (25.5) | 13 (31.7) | ||||
| BRCA1, no. (%) | 2.219 | 0.330 | 0.863 | 0.375 | ||||
| Unmethylated | 504 (82.4) | 256 (84.2) | 217 (81.3) | 31 (75.6) | ||||
| Methylated | 108 (17.6) | 48 (15.8) | 50 (18.7) | 10 (24.4) | ||||
| Presence of hypermethylation in any gene, no. (%) | 8.739 | 0.013* | 8.537 | 0.004* | ||||
| Unmethylated | 70 (11.4) | 24 (7.9) | 42 (15.7) | 4 (9.8) | ||||
| Methylated | 542 (88.6) | 28 (92.1) | 225 (84.3) | 37 (90.2) | ||||
| Mean methylated genes | 3.70 ± 2.33 | 2.83 ± 2.18 | 3.85 ± 2.35 | 21.900 | < 0.001* | −4.431 | < 0.001* | |
| Number of methylated genes, no. (%) | 20.046 | < 0.001* | 16.108 | < 0.001* | ||||
| 0–2 | 254 (41.5) | 108 (35.5) | 135 (50.6) | 11 (26.8) | ||||
| 3–5 | 243 (39.7) | 126 (41.4) | 97 (36.3) | 20 (48.8) | ||||
| 6–10 | 115 (18.8) | 70 (23.0) | 35 (13.1) | 10 (24.4) | ||||
| Prognostic outcomes | ||||||||
| aOverall mortality, no. (%) | 0.059 | 0.011* | 4.547 | 0.033* | ||||
| Survive | 379 (66.4) | 210 (69.1) | 169 (63.3) | 23 (56.1) | ||||
| Death | 192 | 94 (30.9) | 98 (36.7) | 18 (43.9) | ||||
| aCancer-specific mortality, no. (%) | 0.059 | 0.011* | 4.547 | 0.033* | ||||
| Survive | 425 (69.4) | 223 (73.4) | 178 (66.7) | 34 (58.5) | ||||
| Death | 187 (30.6) | 81 (26.6) | 89 (33.3) | 17 (41.5) | ||||
| aIntravesical recurrence, no. (%) | 6.131 | 0.047* | 2.879 | 0.090 | ||||
| No recurrence | 438 (71.6) | 228 (75.0) | 185 (69.3) | 25 (61.0) | ||||
| Recurrence | 174 (28.4) | 76 (25.0) | 82 (30.7) | 16 (39.0) | ||||
| aContralateral recurrence, no. (%) | 6.668 | 0.036* | 0.610 | 0.435 | ||||
| No recurrence | 580 (94.8) | 291 (95.7) | 253 (94.8) | 41 (87.8) | ||||
| Recurrence | 32 (5.2) | 13 (4.3) | 14 (5.2) | 5 (12.2) | ||||
CKD chronic kidney disease, eGFR estimated glomerular filtration rate, SD standard deviation
*Statistically significant
aLog-rank test was used
Discussions
In a meta-analysis which included 17 studies with 12,094 patients, Wu et al. demonstrated that ureteral tumors exhibited worse CSS and recurrence-free survival than renal pelvic tumors based on adjusted HRs; however, no such results were noticed in subgroup analysis of pT3/4 and pN1 tumors, though the authors observed significant heterogeneity among reported articles [4]. The only corresponding study that additionally included molecular work was published in 2013, in which Krabbe et al. found no difference in the expression of p21, p27, p53, cyclin E, and Ki-67 [8].
Regarding the relatively higher stages of renal pelvic tumors, Raman et al. suggested that ureteral tumors tend to be diagnosed earlier due to ureteric obstruction, and thus were likely to be detected at a lower stage [5]. In the current cohort of patients, more patients with renal pelvic tumors were diagnosed due to gross hematuria, while the prevalent presence of hydronephrosis could help the detection of ureteral tumors by ultrasound in annual regular physical examination in many patients.
It’s interesting that the presence of sessile architecture and higher tumor grade was more common in ureteral tumors, which indicated the higher aggressiveness of ureteral tumors, as demonstrated in prior studies [4]. The change of DNA methylation status is regarded to be a key event in transcriptionally repressed regions of the genome [12]. Hypermethylation is a mechanism for repression of gene transcription in cancer [9]. Prior studies on bladder cancer demonstrated aberrant methylation status of some specific gene promoter as a sign of higher aggressiveness and worse prognosis [11, 15–19]. We similarly found that increased number of methylated genes appeared to correlate with worse CSS.
Our results demonstrate that renal pelvic and ureteral tumors, though both belong to UTUC, are not totally biologically homogenous and might behave differently. It’s interesting that the rate of hypermethylation was much more higher in renal pelvis tumors than in the ureter, but the ureteral tumors exhibited higher aggressiveness and relatively worse prognosis. What’s more, it’s notable that on sub-analysis, the number of methylated genes was a stronger driver for oncologic outcomes in ureteral tumors. This being said, however, each gene must also be viewed separately, as the prognostic effect of gene hypermethylation appeared to differ by location, further implicating differences in underlying biology between the two groups.
In a published Meta-analysis ureteral location was related to higher risk of bladder recurrence [21]. Although no statistical difference was found in our study, a more distally located tumor within the ureter could conceivably affect bladder recurrence as seen in our previous publication [22].The analysis with gene methylation status didn’t seem to be very informative for this phenomenon. In a Japanese multi-institutional study, Tanaka et al. found that the patterns of tumor spread was related to primary location of the urothelial carcinoma: patients with ureteral tumors (especially at middle and lower part) tended to suffer from local recurrence in the pelvic cavity, while renal pelvic tumors were associated with higher risk of lung metastasis [7]. The underlying biological mechanisms about the differences in the patterns of tumor metastasis corresponding to tumor location remain to be elucidated in the future.
Our study has several limitations related to the retrospective design, and there might be some selection and recall bias, especially considering some patients were excluded due to the unavailable extracted DNA for test. The exact rate and site of distant metastasis and local recurrence were also incompletely available, which precluded further analysis concerning difference patterns of disease recurrence.
Despite these limitations, our study was the first comparative study that integrated epigenetic information with UTUC tumor location, and to our knowledge, the first study that demonstrated the higher prevalence of gene promoter hyper-methylation in renal pelvic tumors. Indeed, future research is warranted to further elucidate the role that gene methylation plays in the development and biology of renal pelvic and ureteral tumors.
Conclusion
Renal pelvic tumors and ureteral tumors exhibited significant differences in clinicopathologic characteristics and epigenetic biomarkers. Gene promoter methylation might be an important mechanism in explaining distinct tumor patterns and behaviors in UTUC.
Acknowledgements
The author thank the entire staff of Department of Urology, Peking University First Hospital Structured data processing occurred partially using Medbanks’ approach [Medbanks (Beijing) Network Technology CO, Ltd].
Funding
This study was funded by Collaborative Research Foundation of Peking University Health Science Center and National Taiwan University, the College of Medicine (BMU20120318), the Natural Science Foundation of Beijing (7152146), the Clinical Features Research of Capital (No. Z151100004015173), the Capital Health Research and Development of Special (2016–1-4077) and Fund for Fostering Young Scholars of Peking University Health Science Center (BMU2017PY009).
Availability of data and materials
The datasets of the current study are available from the corresponding author on reasonable request.
Abbreviations
- CSS
Cancer specific survival
- CT
Computed tomography
- eGFR
Estimated glomerular filtration rate
- HR
Hazard ratio
- MRI
Magnetic resonance imaging
- MSP
Methylation-sensitive polymerase chain reaction
- OS
Overall survival
- RNU
Radical nephroureterectomy
- UICC
Union for International Cancer Control
- UTUC
Upper tract urothelial carcinoma
- WHO
World Health Organization
Authors’ contributions
DF, SH, XL, LZ (Zhou): Protocol/project development; DF, SH, ZC, GX, LZ (Zhang), XL: Data collection or management; DF, SH GX: Data analysis; DF, SH, NS: Manuscript writing/editing; XL, LZ (Zhou): Critical revision of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study received the ethics approval by the Ethic Committee of Peking University First Hospital (No. 2016–1253).
All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
All patients agreed and signed the informed consent to participate into the study, and they agreed that their information (including clinical information, surgical related data, pathological data and surveillance) would be collected for scientific study and by published in professional medical journals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dong Fang, Email: fdmailbox@126.com.
Shiming He, Email: shiminghe@bjmu.edu.cn.
Gengyan Xiong, Email: xgy13537221@163.com.
Nirmish Singla, Email: nirmish.singla@phhs.org.
Zhenpeng Cao, Email: shenhaiyuanhang@sina.com.
Lei Zhang, Email: zl070501@126.com.
Xuesong Li, Phone: +86-10-83575006, Email: pineneedle@sina.com.
Liqun Zhou, Phone: +86-10-83575006, Email: zhoulqmail@sina.com.
References
- 1.Perez-Utrilla Perez M, Aguilera Bazan A, Alonso Dorrego JM, Viton Herrero R, Cisneros Ledo J, de la Pena Barthel J. Simultaneous cystectomy and Nephroureterectomy due to synchronous upper urinary tract tumors and invasive bladder Cancer: open and laparoscopic approaches. Currt Urol. 2012;6(2):76–81. doi: 10.1159/000343514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Cummings KB. Nephroureterectomy: rationale in the management of transitional cell carcinoma of the upper urinary tract. Urol Clin North Am. 1980;7(3):569–578. [PubMed] [Google Scholar]
- 4.Wu Y, Dong Q, Liu L, Han P, Wei Q. The impact of tumor location and multifocality on prognosis for patients with upper tract urothelial carcinoma: a meta-analysis. Sci Rep. 2014;4:6361. doi: 10.1038/srep06361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raman JD, Ng CK, Scherr DS, Margulis V, Lotan Y, Bensalah K, Patard JJ, Kikuchi E, Montorsi F, Zigeuner R, Weizer A, Bolenz C, Koppie TM, Isbarn H, Jeldres C, Kabbani W, Remzi M, Waldert M, Wood CG, Roscigno M, Oya M, Langner C, Wolf JS, Strobel P, Fernandez M, Karakiewcz P, Shariat SF. Impact of tumor location on prognosis for patients with upper tract urothelial carcinoma managed by radical nephroureterectomy. Eur Urol. 2010;57(6):1072–1079. doi: 10.1016/j.eururo.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Isbarn H, Jeldres C, Shariat SF, Liberman D, Sun M, Lughezzani G, Widmer H, Arjane P, Pharand D, Fisch M, Graefen M, Montorsi F, Perrotte P, Karakiewicz PI. Location of the primary tumor is not an independent predictor of cancer specific mortality in patients with upper urinary tract urothelial carcinoma. J Urol. 2009;182(5):2177–2181. doi: 10.1016/j.juro.2009.07.035. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka N, Kikuchi E, Kanao K, Matsumoto K, Kobayashi H, Ide H, Miyazaki Y, Obata J, Hoshino K, Shirotake S, Akita H, Kosaka T, Miyajima A, Momma T, Nakagawa K, Hasegawa S, Nakajima Y, Jinzaki M, Oya M. Metastatic behavior of upper tract urothelial carcinoma after radical nephroureterectomy: association with primary tumor location. Ann Surg Oncol. 2014;21(3):1038–1045. doi: 10.1245/s10434-013-3349-z. [DOI] [PubMed] [Google Scholar]
- 8.Krabbe LM, Bagrodia A, Westerman ME, Gayed BA, Haddad AQ, Sagalowsky AI, Shariat SF, Kapur P, Lotan Y, Margulis V. Molecular profile of urothelial carcinoma of the upper urinary tract: are pelvicalyceal and ureteral tumors different? World J Urol. 2016;34(1):105–112. doi: 10.1007/s00345-015-1584-6. [DOI] [PubMed] [Google Scholar]
- 9.Yates DR, Catto JW. Distinct patterns and behaviour of urothelial carcinoma with respect to anatomical location: how molecular biomarkers can augment clinico-pathological predictors in upper urinary tract tumours. World J Urol. 2013;31(1):21–29. doi: 10.1007/s00345-012-0946-6. [DOI] [PubMed] [Google Scholar]
- 10.Catto JW, Azzouzi AR, Amira N, Rehman I, Feeley KM, Cross SS, Fromont G, Sibony M, Hamdy FC, Cussenot O, Meuth M. Distinct patterns of microsatellite instability are seen in tumours of the urinary tract. Oncogene. 2003;22(54):8699–8706. doi: 10.1038/sj.onc.1206964. [DOI] [PubMed] [Google Scholar]
- 11.Catto JW, Azzouzi AR, Rehman I, Feeley KM, Cross SS, Amira N, Fromont G, Sibony M, Cussenot O, Meuth M, Hamdy FC. Promoter hypermethylation is associated with tumor location, stage, and subsequent progression in transitional cell carcinoma. J Clin Oncol. 2005;23(13):2903–2910. doi: 10.1200/JCO.2005.03.163. [DOI] [PubMed] [Google Scholar]
- 12.Xiong G, Liu J, Tang Q, Fan Y, Fang D, Yang K, Xie F, Zhang M, Zhang L, Liu L, Zhang C, Yao L, Yang L, Ci W, Zhao W, Gong Y, He Q, Gong K, He Z, Wang G, Li X, Guo Y, Zhou L. Prognostic and predictive value of epigenetic biomarkers and clinical factors in upper tract urothelial carcinoma. Epigenomics. 2015;7(5):733–744. doi: 10.2217/epi.15.34. [DOI] [PubMed] [Google Scholar]
- 13.Ma YC, Zuo L, Chen JH, Luo Q, Yu XQ, Li Y, Xu JS, Huang SM, Wang LN, Huang W, Wang M, Xu GB, Wang HY. Modified glomerular filtration rate estimating equation for Chinese patients with chronic kidney disease. J Am Soc Nephrol. 2006;17(10):2937–2944. doi: 10.1681/ASN.2006040368. [DOI] [PubMed] [Google Scholar]
- 14.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, Zhu T, Wang Z, Zhang H, Qian Z, Xu H, Gao B, Wang W, Gu L, Meng J, Wang J, Feng X, Li Y, Yao X, Zhu J. A novel set of DNA methylation markers in urine sediments for sensitive/specific detection of bladder cancer. Clin Cancer Res. 2007;13(24):7296–7304. doi: 10.1158/1078-0432.CCR-07-0861. [DOI] [PubMed] [Google Scholar]
- 16.Maruyama R, Toyooka S, Toyooka KO, Harada K, Virmani AK, Zochbauer-Muller S, Farinas AJ, Vakar-Lopez F, Minna JD, Sagalowsky A, Czerniak B, Gazdar AF. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 2001;61(24):8659–8663. [PubMed] [Google Scholar]
- 17.Costa VL, Henrique R, Danielsen SA, Duarte-Pereira S, Eknaes M, Skotheim RI, Rodrigues A, Magalhaes JS, Oliveira J, Lothe RA, Teixeira MR, Jeronimo C, Lind GE. Three epigenetic biomarkers, GDF15, TMEFF2, and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin Cancer Res. 2010;16(23):5842–5851. doi: 10.1158/1078-0432.CCR-10-1312. [DOI] [PubMed] [Google Scholar]
- 18.Lee MG, Kim HY, Byun DS, Lee SJ, Lee CH, Kim JI, Chang SG, Chi SG. Frequent epigenetic inactivation of RASSF1A in human bladder carcinoma. Cancer Res. 2001;61(18):6688–6692. [PubMed] [Google Scholar]
- 19.Casadio V, Molinari C, Calistri D, Tebaldi M, Gunelli R, Serra L, Falcini F, Zingaretti C, Silvestrini R, Amadori D, Zoli W. DNA Methylation profiles as predictors of recurrence in non muscle invasive bladder cancer: an MS-MLPA approach. J Exp Clin Cancer Res. 2013;32:94. doi: 10.1186/1756-9966-32-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang GH, Shim YH, Jung HY, Kim WH, Ro JY, Rhyu MG. CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res. 2001;61(7):2847–2851. [PubMed] [Google Scholar]
- 21.Seisen T, Granger B, Colin P, Leon P, Utard G, Renard-Penna R, Comperat E, Mozer P, Cussenot O, Shariat SF, Roupret M. A systematic review and meta-analysis of Clinicopathologic factors linked to Intravesical recurrence after radical Nephroureterectomy to treat upper tract urothelial carcinoma. Eur Urol. 2015;67(6):1122–1133. doi: 10.1016/j.eururo.2014.11.035. [DOI] [PubMed] [Google Scholar]
- 22.Fang D, Xiong GY, Li XS, Chen XP, Zhang L, Yao L, He ZS, Zhou LQ. Pattern and risk factors of intravesical recurrence after nephroureterectomy for upper tract urothelial carcinoma: a large Chinese center experience. J Formos Med Assoc. 2014;113(11):820–827. doi: 10.1016/j.jfma.2013.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets of the current study are available from the corresponding author on reasonable request.


