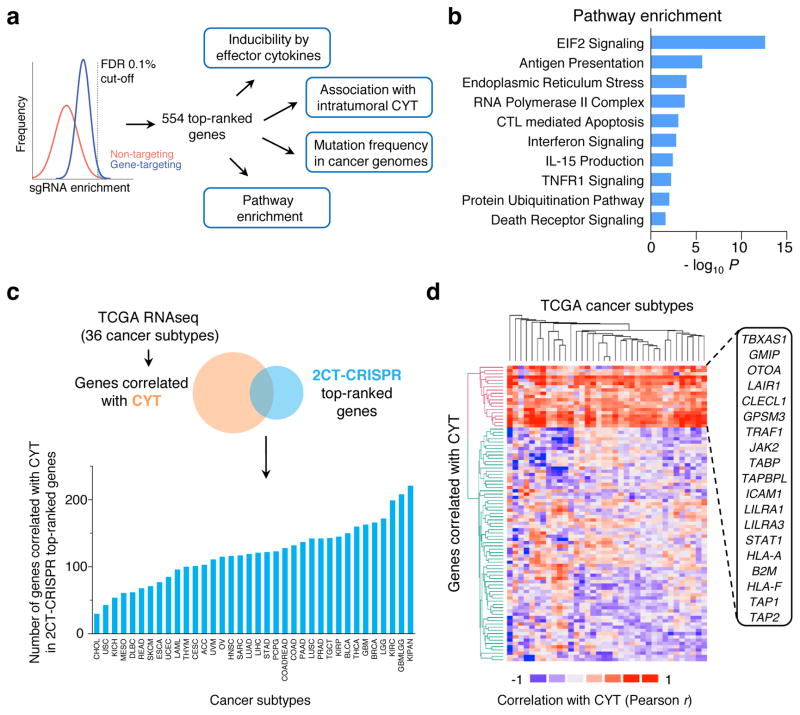
Figure 3. Categorization of candidate essential genes for EFT using available knowledge database.
a, Candidate genes based on FDR cut-off of 0.1% (n = 554 genes) were functionally categorized by multiple methods to understand the biological significance for EFT. b, Most significant pathways by Ingenuity Pathway Analysis (P < 0.05) from top 2CT-CRISPR candidates. c–d, Association of top 554 candidate genes with intratumoral cytolytic activity (CYT, expression of perforin and granzyme) using TCGA datasets. c, RNA-sequencing data for 36 human cancers were obtained from the TCGA database and genes positively correlated with CYT (P < 0.05, Pearson correlation) were intersected with 2CT-CRISPR candidates to quantify number of overlapping genes in each cancer subtype. d, Heatmap showing the partitioning of the clusters of genes based on Pearson correlation coefficient values of CRISPR screen hits with CYT using pan-cancer TCGA data. Individual cancer and pathway heatmaps are available at https://bioinformatics.cancer.gov/publications/restifo.