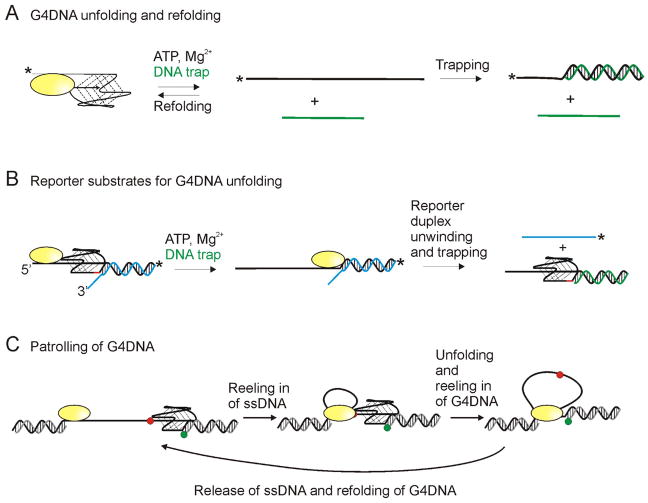
Figure 4. Substrates utilized for G4DNA unfolding studies.
(A) Some experiments for measuring G4DNA unfolding rely on trapping the unfolded duplex as dsDNA to prevent refolding of the G4DNA structure. Experiments typically start with the enzyme prebound to the G4DNA substrate and unfolding is initiated by addition of ATP, Mg2+, and a DNA trap (green) complementary to the G4DNA. G4DNA substrate and dsDNA product can be separated by electrophoresis. * indicates either a fluorescent or radiolabel. Since refolding is a unimolecular process and trapping is a bimolecular process, trapping can be inefficient. (B) Alternatively, G4DNA unfolding can be monitored by measuring the unwinding of a reporter duplex following the G4DNA. Appearance of ssDNA provides a “report” on the unfolding of the G4DNA. A gap of 2 nucleotides (red) between the G4DNA and reporter duplex is necessary for folding of both the G4DNA and dsDNA to occur [83]. (C) Based on smFRET studies, some helicases have been reported to remain anchored at a ssDNA/dsDNA junction while repetitively reeling in the ssDNA and unfolded G4DNA. Green and red dots represent Cy3 and Cy5.