Fig. 3.
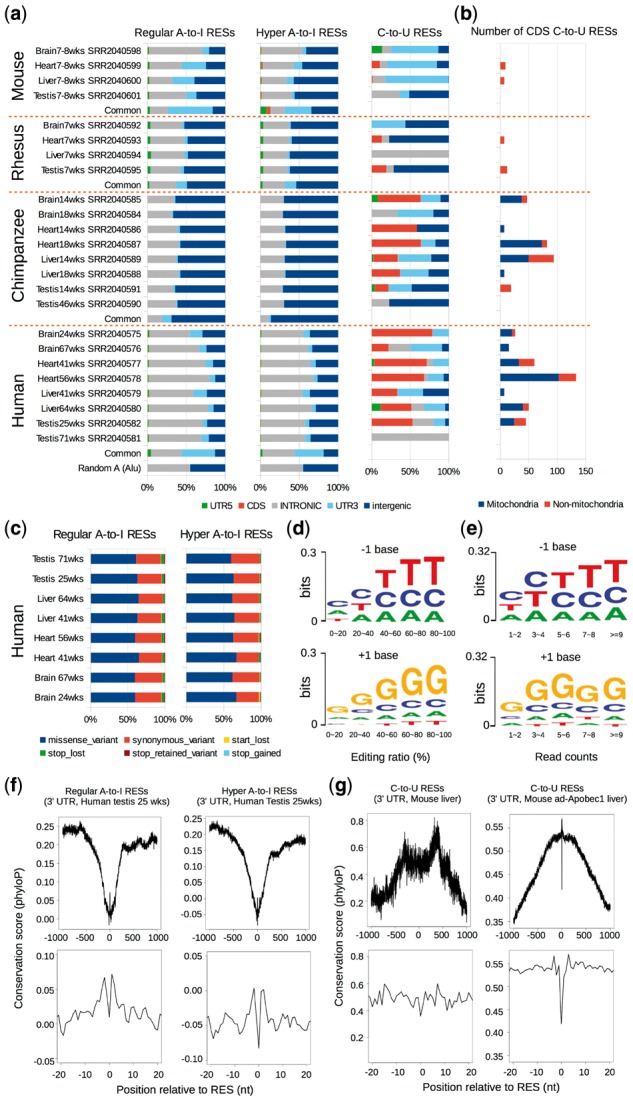
RNA editing in different tissues of human, chimpanzee, rhesus and mouse. (a) The proportions of different categories of regular A-to-I RESs (left), hyper A-to-I RESs (middle) and C-to-U RESs (right) called by SPRINT in different tissues of human, chimpanzee, rhesus and mouse. ‘wks’ refers to ‘weeks’. (b) The number of CDS C-to-U RESs detected in different tissues of the four species. (c) The fractions of potential coding consequences (e.g. missense, synonymous, etc.) of CDS A-to-I RESs called by SPRINT in human. We used Variant Effect Predictor (VEP, http://www.ensembl.org/info/docs/tools/vep/) to annotate the potential coding consequences of human CDS regular and hyper A-to-I RESs. (d) The preference for the nucleotide before (−1, upper) and after (+1, lower) a regular A-to-I RES for RESs with different editing ratios. Here, the read depth of a RES is required to be greater than or equal to five. The nucleotide preference is plotted using WebLogo(Crooks et al., 2004). (e) Similar to (d) except that RESs with different read counts are investigated. (f) The averaged phyloP conservation scores (Pollard et al., 2010) at different positions relative to a regular A-to-I RES (left) or a hyper A-to-I RES (right) in 3’ UTR, with the position ranging from −1000 to +1000 (upper) and from -20 to +20 (lower). (The sequence patterns of A-to-I RESs in other categories can be found in Supplementary Fig. S7). In (d–f), the A-to-I RESs used are from human testis (25 weeks), because human testis has the most number of detected A-to-I RESs among all tissues investigated in this study. (g) Similar to (f), except that C-to-U RESs are investigated. The left and right sub-figures are plotted using the C-to-U RESs identified from mouse liver (7–8 weeks) and mouse ad-Apobec1 liver, respectively
