Fig. 2.
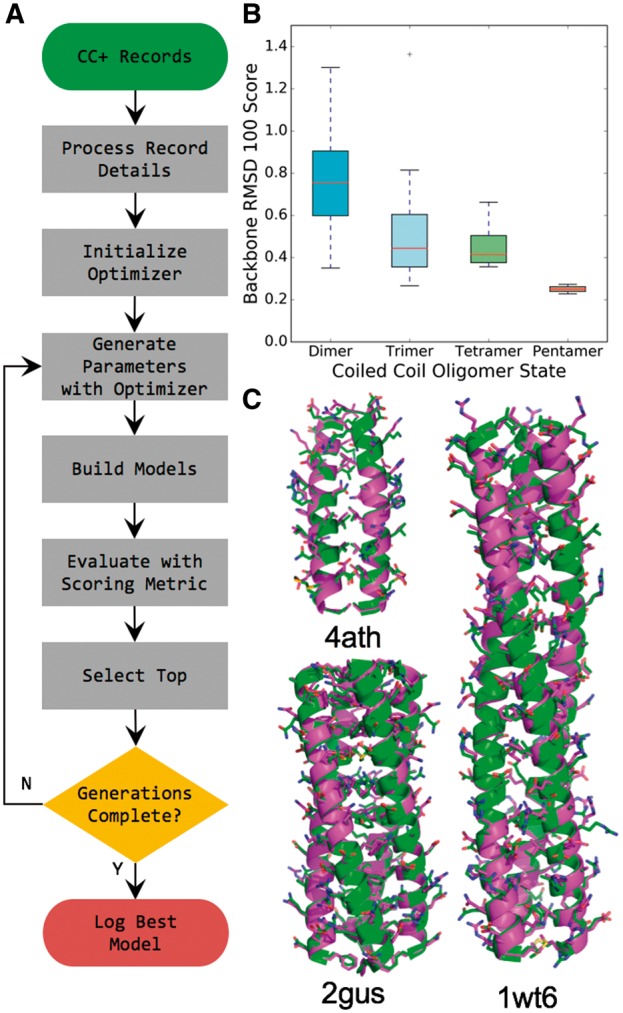
Crystal structures of coiled coils are recreated using parametric model building in ISAMBARD. (A) Model-building methodology for coiled coils employed to test the accuracy of ISAMBARD. The differential evolution optimizer was used with RMSD between the model and the experimental X-ray crystal structure as the scoring metric. (B) Box and whiskers plot of RMSD100 scores for non-redundant, dimers (cyan, n = 66), trimers (light blue, n = 41), tetramers (light green, n = 4) and pentamers (tan, n = 2) in CC+ (Testa et al. 2009), with more than a total of 44 residues. (C) Overlay of experimentally determined structure (green) with corresponding model (magenta), for a dimeric (4ath, RMSD = 0.48 Å), trimeric (1wt6, RMSD = 0.67 Å) and tetrameric (2gus, RMSD = 0.45 Å) coiled coil (Color version of this figure is available at Bioinformatics online.)
