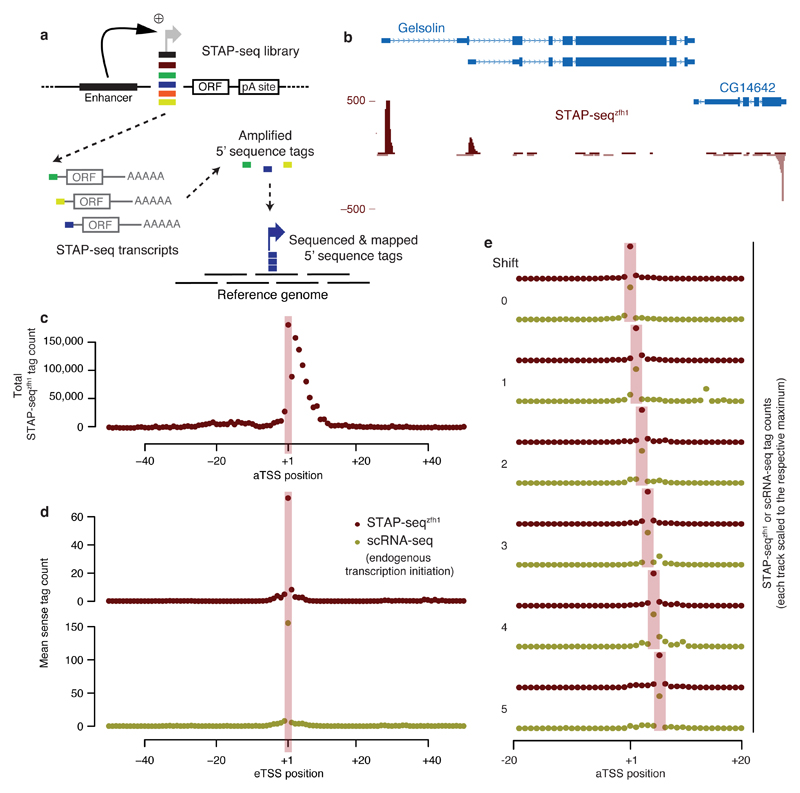
Figure 1. STAP-seq identifies position and orientation of transcription initiation within arbitrary candidate fragments.
(a) Experimental setup of STAP-seq. Short candidate DNA fragments are cloned into a reporter construct that provides an enhancer and a reporter gene (short open reading frame (ORF)). Active candidates initiate reporter transcripts that start with sequence tags depicting the exact TSS. These tags are then sequenced and mapped to the reference genome. (b) UCSC Genome browser screenshot depicting STAP-seq using the zfh1 enhancer. Tag coverage is shown in a strand-specific manner. (c) Cumulative STAP-seqzfh1 tag counts around FlyBase-annotated TSSs (aTSS). (d) Metagene profile of STAP-seqzfh1 tag counts and short-nuclear-capped-RNA-seq (scRNA-seq)18 signals at experimentally determined STAP-seq TSSs (eTSSs). (e) Agreement of STAP-seqzfh1 and scRNA-seq18 for eTSSs that are shifted with respect to aTSSs by 1–5 nt.